EMBRYOPSIDA Pirani & Prado
Gametophyte dominant, independent, multicellular, initially ±globular, not motile, branched; showing gravitropism; glycolate oxidase +, glycolate metabolism in leaf peroxisomes [glyoxysomes], acquisition of phenylalanine lysase* [PAL], flavonoid synthesis*, microbial terpene synthase-like genes +, triterpenoids produced by CYP716 enzymes, CYP73 and phenylpropanoid metabolism [development of phenolic network], xyloglucans in primary cell wall, side chains charged; plant poikilohydrous [protoplasm dessication tolerant], ectohydrous [free water outside plant physiologically important]; thalloid, leafy, with single-celled apical meristem, tissues little differentiated, rhizoids +, unicellular; chloroplasts several per cell, pyrenoids 0; centrioles/centrosomes in vegetative cells 0, microtubules with γ-tubulin along their lengths [?here], interphase microtubules form hoop-like system; metaphase spindle anastral, predictive preprophase band + [with microtubules and F-actin; where new cell wall will form], phragmoplast + [cell wall deposition centrifugal, from around the anaphase spindle], plasmodesmata +; antheridia and archegonia +, jacketed*, surficial; blepharoplast +, centrioles develop de novo, bicentriole pair coaxial, separate at midpoint, centrioles rotate, associated with basal bodies of cilia, multilayered structure + [4 layers: L1, L4, tubules; L2, L3, short vertical lamellae] (0), spline + [tubules from L1 encircling spermatid], basal body 200-250 nm long, associated with amorphous electron-dense material, microtubules in basal end lacking symmetry, stellate array of filaments in transition zone extended, axonemal cap 0 [microtubules disorganized at apex of cilium]; male gametes [spermatozoids] with a left-handed coil, cilia 2, lateral, asymmetrical; oogamy; sporophyte +*, multicellular, growth 3-dimensional*, cuticle +*, plane of first cell division transverse [with respect to long axis of archegonium/embryo sac], sporangium and upper part of seta developing from epibasal cell [towards the archegonial neck, exoscopic], with at least transient apical cell [?level], initially surrounded by and dependent on gametophyte, placental transfer cells +, in both sporophyte and gametophyte, wall ingrowths develop early; suspensor/foot +, cells at foot tip somewhat haustorial; sporangium +, single, terminal, dehiscence longitudinal; meiosis sporic, monoplastidic, MTOC [= MicroTubule Organizing Centre] associated with plastid, sporocytes 4-lobed, cytokinesis simultaneous, preceding nuclear division, quadripolar microtubule system +; wall development both centripetal and centrifugal, 1000 spores/sporangium, sporopollenin in the spore wall* laid down in association with trilamellar layers [white-line centred lamellae; tripartite lamellae]; plastid transmission maternal; nuclear genome [1C] <1.4 pg, main telomere sequence motif TTTAGGG, KNOX1 and KNOX2 [duplication] and LEAFY genes present, ethylene involved in cell elongation; chloroplast genome with close association between trnLUAA and trnFGAA genes [precursors for starch synthesis], tufA, minD, minE genes moved to nucleus; mitochondrial trnS(gcu) and trnN(guu) genes +.
Many of the bolded characters in the characterization above are apomorphies of more or less inclusive clades of streptophytes along the lineage leading to the embryophytes, not apomorphies of crown-group embryophytes per se.
All groups below are crown groups, nearly all are extant. Characters mentioned are those of the immediate common ancestor of the group, [] contains explanatory material, () features common in clade, exact status unclear.
POLYSPORANGIOPHYTA†
Sporophyte well developed, branched, branching dichotomous, potentially indeterminate; hydroids +; stomata on stem; sporangia several, terminal; spore walls not multilamellate [?here].
II. TRACHEOPHYTA / VASCULAR PLANTS
Sporophyte long lived, cells polyplastidic, photosynthetic red light response, stomata open in response to blue light; plant homoiohydrous [water content of protoplasm relatively stable]; control of leaf hydration passive; plant endohydrous [physiologically important free water inside plant]; PIN[auxin efflux facilitators]-mediated polar auxin transport; (condensed or nonhydrolyzable tannins/proanthocyanidins +); borate cross-linked rhamnogalactan II, xyloglucans with side chains uncharged [?level], in secondary walls of vascular and mechanical tissue; lignins +; roots +, often ≤1 mm across, root hairs and root cap +; stem apex multicellular [several apical initials, no tunica], with cytohistochemical zonation, plasmodesmata formation based on cell lineage; vascular development acropetal, tracheids +, in both protoxylem and metaxylem, G- and S-types; sieve cells + [nucleus degenerating]; endodermis +; stomata numerous, involved in gas exchange; leaves +, vascularized, spirally arranged, blades with mean venation density ca 1.8 mm/mm2 [to 5 mm/mm2], all epidermal cells with chloroplasts; sporangia in strobili, sporangia adaxial, columella 0; tapetum glandular; sporophyte-gametophyte junction lacking dead gametophytic cells, mucilage, ?position of transfer cells; MTOCs not associated with plastids, basal body 350-550 nm long, stellate array in transition region initially joining microtubule triplets; archegonia embedded/sunken [only neck protruding]; embryo suspensor +, shoot apex developing away from micropyle/archegonial neck [from hypobasal cell, endoscopic], root lateral with respect to the longitudinal axis of the embryo [plant homorhizic].
[MONILOPHYTA + LIGNOPHYTA]Sporophyte growth ± monopodial, branching spiral; roots endomycorrhizal [with Glomeromycota], lateral roots +, endogenous; G-type tracheids +, with scalariform-bordered pits; leaves with apical/marginal growth, venation development basipetal, growth determinate; sporangium dehiscence by a single longitudinal slit; cells polyplastidic, MTOCs diffuse, perinuclear, migratory; blepharoplasts +, paired, with electron-dense material, centrioles on periphery, male gametes multiciliate; nuclear genome [1C] 7.6-10 pg [mode]; chloroplast long single copy ca 30kb inversion [from psbM to ycf2]; mitochondrion with loss of 4 genes, absence of numerous group II introns; LITTLE ZIPPER proteins.
LIGNOPHYTA†
Sporophyte woody; stem branching axillary, buds exogenous; lateral root origin from the pericycle; cork cambium + [producing cork abaxially], vascular cambium bifacial [producing phloem abaxially and xylem adaxially].
SEED PLANTS† / SPERMATOPHYTA†
Growth of plant bipolar [plumule/stem and radicle/root independent, roots positively geotropic]; plants heterosporous; megasporangium surrounded by cupule [i.e. = unitegmic ovule, cupule = integument]; pollen lands on ovule; megaspore germination endosporic, female gametophyte initially retained on the plant, free-nuclear/syncytial to start with, walls then coming to surround the individual nuclei, process proceeding centripetally.
EXTANT SEED PLANTS
Plant evergreen; nicotinic acid metabolised to trigonelline, (cyanogenesis via tyrosine pathway); microbial terpene synthase-like genes 0; primary cell walls rich in xyloglucans and/or glucomannans, 25-30% pectin [Type I walls]; lignin chains started by monolignol dimerization [resinols common], particularly with guaiacyl and p-hydroxyphenyl [G + H] units [sinapyl units uncommon, no Maüle reaction]; roots often ≥1 mm across, stele diarch to pentarch, xylem and phloem originating on alternating radii, cork cambium deep seated, gravitropism response fast; stem apical meristem complex [with quiescent centre, etc.], plasmodesma density in SAM 1.6-6.2[mean]/μm2 [interface-specific plasmodesmatal network]; eustele +, protoxylem endarch, endodermis 0; wood homoxylous, tracheids and rays alone, tracheid/tracheid pits circular, bordered; mature sieve tube/cell lacking functioning nucleus, sieve tube plastids with starch grains; phloem fibres +; cork cambium superficial; leaf nodes 1:1, a single trace leaving the vascular sympodium; leaf vascular bundles amphicribral; guard cells the only epidermal cells with chloroplasts, stomatal pore with active opening in response to leaf hydration, control by abscisic acid, metabolic regulation of water use efficiency, etc.; branching by axillary buds, exogenous; prophylls two, lateral; leaves with petiole and lamina, development basipetal, lamina simple; sporangia borne on sporophylls; spores not dormant; microsporophylls aggregated in indeterminate cones/strobili; grains monosulcate, aperture in ana- position [distal], primexine + [involved in exine pattern formation with deposition of sporopollenin from tapetum there], exine and intine homogeneous, exine alveolar/honeycomb; ovules with parietal tissue [= crassinucellate], megaspore tetrad linear, functional megaspore single, chalazal, sporopollenin 0; gametophyte ± wholly dependent on sporophyte, development initially endosporic [apical cell 0, rhizoids 0, etc.]; male gametophyte with tube developing from distal end of grain, male gametes two, developing after pollination, with cell walls; embryo cellular ab initio, suspensor short-minute, embryonic axis straight [shoot and root at opposite ends], primary root/radicle produces taproot [= allorhizic], cotyledons 2; embryo ± dormant; chloroplast ycf2 gene in inverted repeat, trans splicing of five mitochondrial group II introns, rpl6 gene absent; ??whole nuclear genome duplication [ζ/zeta duplication event], 2C genome size (0.71-)1.99(-5.49) pg, two copies of LEAFY gene, PHY gene duplications [three - [BP [A/N + C/O]] - copies], 5.8S and 5S rDNA in separate clusters.
IID. ANGIOSPERMAE / MAGNOLIOPHYTA
Lignans, O-methyl flavonols, dihydroflavonols, triterpenoid oleanane, apigenin and/or luteolin scattered, [cyanogenesis in ANA grade?], lignin also with syringyl units common [G + S lignin, positive Maüle reaction - syringyl:guaiacyl ratio more than 2-2.5:1], hemicelluloses as xyloglucans; root cap meristem closed (open); pith relatively inconspicuous, lateral roots initiated immediately to the side of [when diarch] or opposite xylem poles; epidermis probably originating from inner layer of root cap, trichoblasts [differentiated root hair-forming cells] 0, hypodermis suberised and with Casparian strip [= exodermis]; shoot apex with tunica-corpus construction, tunica 2-layered; starch grains simple; primary cell wall mostly with pectic polysaccharides, poor in mannans; tracheid:tracheid [end wall] plates with scalariform pitting, multiseriate rays +, wood parenchyma +; sieve tubes enucleate, sieve plates with pores (0.1-)0.5-10< µm across, cytoplasm with P-proteins, not occluding pores of plate, companion cell and sieve tube from same mother cell; ?phloem loading/sugar transport; nodes 1:?; dark reversal Pfr → Pr; protoplasm dessication tolerant [plant poikilohydric]; stomata randomly oriented, brachyparacytic [ends of subsidiary cells ± level with ends of guard cells], outer stomatal ledges producing vestibule, reduction in stomatal conductance with increasing CO2 concentration; lamina formed from the primordial leaf apex, margins toothed, development of venation acropetal, overall growth ± diffuse, secondary veins pinnate, fine venation hierarchical-reticulate, (1.7-)4.1(-5.7) mm/mm2, vein endings free; flowers perfect, pedicellate, ± haplomorphic, protogynous; parts free, numbers variable, development centripetal; P = T, petal-like, each with a single trace, outer members not sharply differentiated from the others, not enclosing the floral bud; A many, filament not sharply distinguished from anther, stout, broad, with a single trace, anther introrse, tetrasporangiate, sporangia in two groups of two [dithecal], each theca dehiscing longitudinally by a common slit, ± embedded in the filament, walls with at least outer secondary parietal cells dividing, endothecium +, cells elongated at right angles to long axis of anther; tapetal cells binucleate; microspore mother cells in a block, microsporogenesis successive, walls developing by centripetal furrowing; pollen subspherical, tectum continuous or microperforate, ektexine columellate, endexine restricted to the apertural regions, thin, compact, intine in apertural areas thick, orbicules +, pollenkitt +; nectary 0; carpels present, superior, free, several, spiral, ascidiate [postgenital occlusion by secretion], stylulus at most short [shorter than ovary], hollow, cavity not lined by distinct epidermal layer, stigma ± decurrent, carinal, dry; suprastylar extragynoecial compitum +; ovules few [?1]/carpel, marginal, anatropous, bitegmic, micropyle endostomal, outer integument 2-3 cells across, often largely subdermal in origin, inner integument 2-3 cells across, often dermal in origin, parietal tissue 1-3 cells across, nucellar cap?; megasporocyte single, hypodermal, functional megaspore lacking cuticle; female gametophyte lacking chlorophyll, four-celled [one module, egg and polar nuclei sisters]; ovule not increasing in size between pollination and fertilization; pollen grains bicellular at dispersal, germinating in less than 3 hours, siphonogamy, pollen tube unbranched, growing towards the ovule, between cells, growth rate (ca 10-)80-20,000 µm h-1, tube apex of pectins, wall with callose, lumen with callose plugs, penetration of ovules via micropyle [porogamous], whole process takes ca 18 hours, distance to first ovule 1.1-2.1 mm; male gametophytes tricellular, gametes 2, lacking cell walls, ciliae 0, double fertilization +, ovules aborting unless fertilized; fruit indehiscent, P deciduous; mature seed much larger than fertilized ovule, small [<5 mm long], dry [no sarcotesta], exotestal; endosperm +, ?diploid [one polar nucleus + male gamete], cellular, development heteropolar [first division oblique, micropylar end initially with a single large cell, divisions uniseriate, chalazal cell smaller, divisions in several planes], copious, oily and/or proteinaceous, embryo short [<¼ length of seed]; plastid and mitochondrial transmission maternal; Arabidopsis-type telomeres [(TTTAGGG)n]; nuclear genome [2C] (0.57-)1.45(-3.71) [1 pg = 109 base pairs], ??whole nuclear genome duplication [ε/epsilon event]; ndhB gene 21 codons enlarged at the 5' end, single copy of LEAFY and RPB2 gene, knox genes extensively duplicated [A1-A4], AP1/FUL gene, palaeo AP3 and PI genes [paralogous B-class genes] +, with "DEAER" motif, SEP3/LOFSEP and three copies of the PHY gene, [PHYB [PHYA + PHYC]]; chloroplast IR expansions, chlB, -L, -N, trnP-GGG genes 0.
[NYMPHAEALES [AUSTROBAILEYALES [MONOCOTS [[CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]]]]: wood fibres +; axial parenchyma diffuse or diffuse-in-aggregates; pollen monosulcate [anasulcate], tectum reticulate-perforate [here?]; ?genome duplication; "DEAER" motif in AP3 and PI genes lost, gaps in these genes.
[AUSTROBAILEYALES [MONOCOTS [[CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]]]: phloem loading passive, via symplast, plasmodesmata numerous; vessel elements with scalariform perforation plates in primary xylem; essential oils in specialized cells [lamina and P ± pellucid-punctate]; tension wood + [reaction wood: with gelatinous fibres, G-fibres, on adaxial side of branch/stem junction]; anther wall with outer secondary parietal cell layer dividing; tectum reticulate; nucellar cap + [character lost where in eudicots?]; 12BP [4 amino acids] deletion in P1 gene.
[MONOCOTS [CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]] / MESANGIOSPERMAE: sesquiterpene synthase subfamily a [TPS-a] [?level], polyacetate derived anthraquinones + [?level]; outer epidermal walls of root elongation zone with cellulose fibrils oriented transverse to root axis; P more or less whorled, 3-merous [?here]; pollen tube growth intra-gynoecial; extragynoecial compitum 0; carpels plicate [?here]; embryo sac monosporic [spore chalazal], 8-celled, bipolar [Polygonum type], antipodal cells persisting; endosperm triploid. - Back to Main Tree
Note: In all node characterizations, boldface denotes a possible apomorphy, (....) denotes a feature the exact status of which in the clade is uncertain, [....] includes explanatory material; other text lists features found pretty much throughout the clade. However, the precise node to which many characters, particularly the more cryptic ones, should be assigned is unclear. This is partly because homoplasy is very common, in addition, basic information for all too many characters is very incomplete, frequently coming from taxa well embedded in the clade of interest and so making the position of any putative apomorphy uncertain. Then there are the not-so-trivial issues of how character states are delimited and ancestral states are reconstructed (see above).
Age. N.B.: Ages in this paragraph are largely based on the topology [MONOCOTS [[CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]]. N. Zhang et al. (2012) suggested an age of (163-)145(-133) Ma, Xue et al. (2012) an age of ca 146.4 Ma, and Naumann et al. (2013) an age of about 148.5 Ma. Some other estimates are older, e.g. 176 Ma (Foster et al. 2016a, q.v. for details). Looking at the pattern of duplication of SEPALLATA genes, and Yockteng et al. (2013) dated this node to around 187-137.4 Ma; (216, 197-)191, 154(-141) and ca 217 Ma are the somewhat older spread of ages on Zheng et al. (2014) and Z. Wu et al. (2014) respectively, and ca 219 Ma in Tank et al. (2015: Table S1) and (225-)198(-173) Ma of Salomo et al. (2017) are also rather older. Bell et al. (2010: Chloranthaceae sister to [Magnoliidae + everything else], but not monocots) suggested ages for this node of (152-)140(-128) or (135-)127(-119) Ma depending on the method used. H.-T. Li et al. (2019) estimate an age of (193-)164(-146) Ma (ages in Moore et al. 2007; S. A. Smith et al. 2010; Clarke et al. 2011; Magallón et al. 2013, 2015; Zanne et al. (2014; Zeng et al. 2014; Foster et al. 2017; Barba-Montoya et al. 2018 - see H.-T. Li et al. 2019: table 2 - are 210-133 Ma).
For the clade [magnoliids [eudicots ...]], no Chloranthaceae, Y. Yang et al. (2020: Suppl. Fig. 22) suggested an age of around 153.6 Ma.
The age of this branching point is estimated at (190-)167(-47) Ma (95% HPD, Smith et al. 2010, Table S3), while a fossil-based estimate is ca 100 Ma (Crepet et al. 2004) [Check] and one based on pollen is somewhat in excess of 163.5 Ma (Zavialova & Tekleva 2021: Fig. 11).
Evolution: Divergence & Distribution. The fact that relationships in this area (see Phylogeny immediately below) are so difficult to work out may be because the divergences between clades here occurred within about 4 Ma (Rendón-Anaya et al. 2019) and ILS and hybridization may have confused the issue.
Tank et al. (2015) suggest that an increase in net diversification at this node might be linked to the ε/epsilon whole genome duplication that characterises all angiosperms; see also P. Soltis and Soltis (2016) for floral evolution here. If [Chloranthales + Ceratophyllales] - or just Chloranthaceae - were sister to all other mesangiosperms (see below), this increase would have to be placed at the node above their place of departure.
For the distribution of isoquinoline alkaloids, also known as 1-benzyltetrahydroisoquinoline or 1-btiq alkaloids, see Waterman (1999, 2007). How to optimise them on the tree is unclear. They are known from Chloranthaceae, the magnoliids, and eudicots, so if there is a clade [[Chloranthaceae + magnoliids] [monocots [Ceratophyllaceae + eudicots]]], as has been recognized here until xii.2021, where they would be optmised on the tree is somewhat unclear. Another distinctive feature, a crystalliferous endotesta, is found in Chloranthaceae, Magnoliaceae and Aristolochiaceae (Friis et al. 2022a, b).
Ecology & Physiology. Primary xylem in which the vessels have simple perforation plates and occur in both protoxylem and metaxylem in the leaves has evolved several times within this clade (Feild & Brodribb 2013); ceteris paribus, such vessels conduct water well. This then allowed the miniaturization of the foliar vein reticulum through the development of veins that are narrow yet still conduct water adequately, in turn allowing dense leaf venation and all that this entails in terms of high rates of photosynthesis and evaporation, etc.; see also elsewhere.
Genes & Genomes. Evans and Rees (1971) discuss variation in the length of the mitotic cycle - that in eudicots is ca 4 hours longer than that in monocots (interphase, G1, is involved, 16 species sampled).
Chemistry, Morphology, etc.. The betalains of core Caryophyllales have biosynthetic similarities with benzylisoquinoline alkaloids. For the sesquiterpene synthase subfamily a, see the Amborella Genome Project (2013); not in Amborellaceae. For the orientation of cellulose fibrils in the outer epidermal walls of root elongation zone, see Kerstens and Verbelen (2002); I do not know what happens in the ANITA grade and in gymnosperms, and magnoliids and eudicots are very poorly sampled. This is perhaps the best place to put triploid endosperm on the tree; the other would be as a synapomorphy for all angiosperms, but in that case it would subsequently be lost twice, or lost once and then regained.
I largely follow Ronse De Craene et al. (2003) for the insertion of floral organs. To add where?: A whorled.
Phylogeny. Relationships between the major clades immediately above the basal pectinations of the angiosperm tree, the ANA grade, i.e., [Amborellales [Nymphaeales [Austrobaileyales [??]]]]), remain unclear (see also Zeng et al. 2014; Wickett et al. 2014 for earlier summaries). Pending further studies, apomorphies for a topology [CERATOPHYLLALES [MONOCOTS [[CHLORANTHALES + MAGNOLIIDS] EUDICOTS]]] are suggested, but the placement of Ceratophyllales in particular is unclear (see also P. Soltis et al. 1999; D. Soltis et al. 2005b; c.f. A.P.G. IV 2016, and in particular A.P.G. III 2009). Relationships between all these groups are discussed immediately below, and for further details of the five clades involved, see eudicots, magnoliids, Ceratophyllales, Chloranthales and monocots.
In the following discussion, the ANA grade is considered fixed, as is the monophyly of the magnoliids, monocots and eudicots, and Chloranthaceae and Ceratophyllaceae. However, monocots in Goremykin et al. (2005) did not always form a monophyletic group, and although this is a rather unlikely result it has been obtained in some studies where the nuclear 18S gene has been included (Troitsky et al. 1991: see Duvall et al. 2006 for references).
There are two related sets of questions to be addressed: First, where in particular are Chloranthales and Ceratophyllum to be placed in the tree?, and second, what are the relationships between the magnoliids, monocots and eudicots? Zeng et al. (2014: suppl. Fig 1) and Y. Yang et al. (2020: Suppl. Fig. 1) outline some fifteen hypotheses of relationships for these groups - not all of them are discussed below. These two sets of questions are directly linked, for example, Graham et al. (2005) found that the inclusion of Ceratophyllum and Chloranthaceae could destabilise relationships among other early branching clades, e.g., the position of the monocots became labile. Over the years, the positions of Ceratophyllaceae and Chloranthaceae have remained particularly uncertain, and Piperales have also tended to wander around the tree. Another in part unrelated issue is the role of aquatics in early angiosperm evolution, and in particular, how diverse and/or abundant were the two early-diverging aquatic clades, Nymphaeales and Ceratophyllaceae and their associates, in the earlier Cretaceous? Aquatic plants play a particularly prominent role in some reconstructions of early angiosperm evolution (see also elsewhere).
As data sets increase in size and sampling of some clades is necessarily constrained - perhaps more Chloranthaceae could be included, but addition of more Ceratophyllaceae is unlikely to affect the issue - the amount of data and the analytical techniques used are becoming increasingly important (e.g. Jarvis et al. 2014). Thus different seed plant topologies were obtained from analyses using single genes or the same number of sites chosen from twelve separate loci (Burleigh & Mathews 2007a); maximum likelihood and maximum parsimony analyses might show systematic error (Burleigh & Mathews 2007b), and so on. Interestingly, when nuclear genomes started to be analysed, we seemed to return to a problem most evident at the beginning of molecular studies - sampling (e.g. Soltis & Soltis 2019), however, the number of nuclear sequences are now increasing by leaps and bounds, however, there may may be analytical problems like the failure to use site-heterogeneous substitution models as suggested by Y. Wang et al. (2024). Below only more frequently-found relationships are discussed, but in comprehensive analyses such as Moore et al. (2010) and Sun et al. (2014) other "minority" relationships can usually be found.
1. Chloranthaceae and Ceratophyllaceae.. A variety of relationships involving the two groups have been suggested.
1A. The two taxa are not immediately related. Hilu et al. (2003: a matK analysis alone), suggested that Ceratophyllaceae were sister to eudicots (see also D. Soltis et al. 2000; Borsch et al. 2005: three rapidly evolving genes, 62% jacknife, 96% posterior probabilities; Müller et al. 2006: 96% posterior probability; some analyses in Saarela et al. 2007; Soltis et al. 2007a: support weak; Qiu & Estabrook 2008; most analyses in Drew et al. 2014; Barba-Montoya et al. 2018; Gitzendanner et al. 2018a: weak support; H.-T. Li et al. 2019, 2021: very extensive plastome analyses, only moderate support; Ramírez-Barahona et al. 2020), and this topology is quite commonly obtained. The clade [Ceratophyllaceae + eudicots] was also recovered both by Givnish et al. (2016b), although Piper sometimes attached itself to Ceratophyllum, and also by Y. Yang et al. (2020: nuclear genomes), although in neither was Chloranthaceae included. Interestingly, in the latter study, in some analyses about one third of the single gene trees supported a relationship [Ceratophyllum + eudicots], a third that of [Ceratophyllum + monocots], and a third that of [Ceratophyllum + magnoliids]... Y. Wang et al. (2024) found quite good support for the relationships [[Monocots + Ceratophyllales] [Magnoliales [Chloranthales + Eudicots]]]. In Zuntini et al. (2024) Ceratophyllum was found to be sister to all angiosperms except the ANA grade, although support was underwhelming, however, this last relationship is followed here. Of course, since Ceratophyllum is a very highly derived aquatic it is difficult to relate its morphology to any of the groups with which it has been associated.
A [Chloranthaceae + magnoliids] clade has quite commonly been recovered, as by Jansen et al. (2006b), Hansen et al. (2007), Ruhfel et al. (2014: whole chloroplast genomes, support not strong), Drew et al. (2014: most analyses), Foster et al. (2016: support not strong, 2017) and Gitzendanner et al. (2018a: chloroplast data, weak support). Ceratophyllum, with quite a long branch, was sister to eudicots (in some reconstructions, Piper, with a very long branch, was also involved), and Chloranthus was sister to the magnoliids (Jansen et al. 2007: moderate to strong support), a position also found by Saarela et al. (2007), many analyses in Moore et al. (2007, 2010), Smith et al. (2010), L. zheng et al. (2014), Magallón et al. (2015), L. Zhang et al. (2019), O.T.P.T.I. (2019) and Zuntini et al. (2024). W. J. Baker et al. (2021a: see Seed Plant Tree) had found the relationships [Chloranthaceae [[Canellales + Piperales (Lactoris moved a few nodes down the tree)] [Laurales + Magnoliales]]] - see also below.
Other suggestions? J. Ma et al. (2021) and X. Guo et al. (2021), published on the relationships between the five mesangiosperm groups following their analyses of the genome of Chloranthus (they looked at C. sessilifolius and C. spicatus respectively); similar amounts of data were produced using the same basic methods (I happened to read Guo et al. first). The genome is largely made up of repetitive elements - 64% in Ma et al. (2021), with long terminal repeats being ca 54.8% of the genome. A sister group relationship between Chloranthaceae (three genera included) and the magnoliids was recovered by Guo et al. (2021), and they noted that the two groups both had many genes with very long introns, more syntenic blocks in common, etc., and such features separated them from monocots, eudicots and Ceratophyllum (see also Lv et al. 2020). They argued strongly for a [Chloranthaceae + magnoliid] grouping, while Ceratophyllum was sister to the eudicots (Guo et al. 2021). Although by no means all single gene analyses recovered the same relationships for Chloranthaceae, over half did, however, nuclear and plastid gene trees were incongruent, in the latter the grouping [monocots [Ceratophyllum + eudicots]] was recovered. Guo et al. (2021: Fig.), noting such conflicting relationships, thought that there had been hybridization between monocots, presumably the stem clade, and something immediately basal to the eudicots. Ma et al. (2021) suggested that there had been hybridizations between monocots and Nymphaea, monocots and magnoliids, and between Chloranthales and the ancestor of [eudicots + Ceratophyllales]; again, there were extensive conflicting/very poorly supported relationships (see also Stull et al. 2023).
L. Yang et al. (2020) looked at 1594 orthologous protein-coding genes in 151 taxa, although only a little over half of these genes were phylogenetically informative. The basic relationships they obtained were ...[Ceratophyllales [monocots [magnoliids [Chloranthales + eudicots]]]] in a concatenation/IQ-TREE analysis and ...[monocots [magnoliids [Ceratophyllales [Chloranthales + eudicots]]]] in a coalescent/ASTRAL analysis; support for the former topology was notably weak, somewhat stronger in the latter. Analyses using datasets of 756 and 296 genes returned the topology ...[monocots [magnoliids [Chloranthales [Ceratophyllales + eudicots]]]] (Yang et al. 2020), with similar somewhat stronger support. A polytomy could not be rejected, and a fully bifurcating tree did not seem to be the best way to representat what was going on.
Rather unusually, Z.-D. Chen et al. (2016) found that Chloranthaceae were sister to the monocots (and Ceratophyllaceae sister to the eudicots) (see also Globoff et al. 2009; Ramírez-Barahona et al. 2020). The relationships [eudicots [Ceratophyllaceae + Piperaceae]] were found by Xue et al. (2012: chloroplast genome ML analysis), although relationships around here were not the focus of the study, nor were they in a PHYC analysis in Hertweck et al. (2015) in which the relationships [Chloranthaceae [[Ceratophyllum + some of the Magnoliales included] [[Acorus + Eudicots] other monocots]]] were obtained.
Drew et al. (2014) also found topologies like [Chloranthaceae [monocots [Ceratophyllaceae + Piperaceae]]], while there was fairly good support for a clade [monocots + Ceratophyllaceae] in a compartmentalised 6-gene analysis (Zanis et al. 2003; see also Y.-L. Qiu et al. 1999; some analyses in Saarela et al. 2007). Qiu et al. (2005; see also Löhne & Borsch 2005) found initial rather strong bootstrap support for a [monocot + Ceratophyllaceae] clade in a 9-gene analysis being vitiated by the failure to obtain much support in any of the subanalyses and by details of the topology obtained in the 9-gene analysis itself (e.g. Acorus sister to the Alismatales included) that are rather improbable. Moore et al. (2010) i.a. found Ceratophyllum, or [Ceratophyllum + Piper] to be sister to monocots and Chloranthaceae sister to a clade including magnoliids, monocots, and eudicots; the latter pair of relationships with moderate jacknife support were found by Davis et al. (2013) in their comprehensive chloroplast analysis focussing on monocots. Indeed, a [Ceratophyllaceae + Piperaceae] clade is not uncommonly found, as by Petersen et al. (2015), and there Chloranthaceae were sister to all angiosperms apart from the ANITA grade (see also P. S. Soltis et al. 2015) - but neither position had much support. Goremykin et al. (2009b: Chloranthaceae not included) found a [Ceratophyllum + magnoliid] clade after removing 2,500 highly variable positions from an analysis of chloroplast genome data; with the removal of 1,000 positions Ceratophyllum was sister to a [monocot + eudicot] clade. Some analyses have suggested a sister taxon relationship between Chloranthaceae and eudicots (Borsch et al. 2003; P. S. Soltis et al. 2015), and this relationship was favoured in several concatenation-based transcriptome analyses by Wickett et al. (2014; no Ceratophyllaceae). Barba-Montoya et al. (2018) placed Chloranthaceae as sister to all angiosperms minus the ANA grade, and this relationship was also found, but with very low support, in the plastome analyses of H.-T. Li et al. (2019, 2021), relationships then being [magnoliids [monocots [Ceratophyllales + eudicots]]], and support was quite high.
Other studies also show no clear pattern (e.g. Jansen et al. 2006a; Qiu et al. 2006b). One alignment of 18S/26S nuclear ribosomal data suggested a clade [Ceratophyllaceae + Tofieldiaceae] embedded within the monocots, but thankfully with little support (Maia et al. 2014), while Du et al. (2016) considered Nymphaeales, Acorales + Alismatales, and Ceratophyllales to be extant members of an early radiation of aquatic angiosperms. Morton (2011) found weak support for Ceratophyllaceae as sister to all other angiosperms, while in the massive parsimony analysis of Goloboff et al. (2009) the relationships [Ceratophyllum [[Amborella + Nymphaeales] all other angiosperms]] were recovered. W. J. Baker et al. (2021a: see Seed Plant Tree) found the rather unexpected relationships [Lactoris [Ceratophyllum [Chloranthaceae [magnoliids]]]] in their preliminary Angiosperms353 genome analysis, however, relationships in subsequent trees have been rather more conventional.
1B. The two are sister taxa. A [Chloranthales + Ceratophyllales] clade is quite often obtained. Analyses in Duvall et al. (2006) and Y.-L. Qiu et al. (2010: chloroplast data, support weak) recovered this clade, as did those in N. Zhang et al. (2012); see also Huang et al. (2010), weak support using the ycf2 gene, Moore et al. (2011), very weak support, and Kvacek et al. (2016: morphological analyses, inc. fossils). In the comprehensive analyses of Sun et al. (2014) this clade was also frequently recovered, although in a variety of positions, while earlier analyses in Mathews (2006a) had suggested an unresolved position somewhere above Austrobaileyales. Indeed, if there is a [Chloranthales + Ceratophyllales] clade, the question is, where is it to be placed? Usually it is associated with magnoliids, monocots or core eudicots in particular (so see below), but sometimes it is placed sister to all other angiosperms except the ANITA grade (e.g. Endress & Doyle 2009; Doyle & Endress 2010; Doyle & Upchurch 2014; Tekleva et al. 2021 and references).
Members of the two groups have fascinating morphologies, both being very reduced florally, and analyses of morphological features may suggest a relationship between the two. Thus the "flower" of Ceratophyllum with 3 to many stamens is perhaps best treated as an inflorescence made up of flowers that consist of a single stamen and nothing more, and this has major effects on the scoring of several characters (e.g. Endress 1994d; Endress & Doyle 2015). Analyses of morphological data gave a [Ceratophyllum + Chloranthaceae] clade with quite strong support (Endress & Doyle 2009). Kvacek et al. (2012) linked Pseudoasterophyllites, ca 97 Ma (Cenomanian) and vegetatively somewhat like Ceratophyllum, with Tucanopollis, an abundant palynomorph from Africa-South America over 125 Ma, and Pseudoasterophyllites tends to link Chloranthaceae and Ceratophyllum (Doyle et al. 2015; Doyle & Endress 2018), and perhaps also Appomattoxia, another fossil (Kvacek et al. 2016). Similarly, Montsechia (= Montsechiaceae), fairly recently descrtibed and variously identified, is either Ceratophyllales or to be placed in a [Chloranthaceae + Ceratophyllum] clade (Gomez et al. 2015); indeed, all these fossils (Tucanopollis was not included, Appomattoxia was) linked with Ceratophyllceae rather than Chloranthaceae in later analyses (Gomez et al. 2020). The fossil chloranthalean Canrightiopsis goes with Ceratophyllum - and with the odd magnolialian plant - in a splits-graph analysis (Friis et al. 2015a). If the staminate flower of Ceratophyllum is interpreted as being simply a single stamen, then much of this reduction must have occurred in the common ancestor of Chloranthaceae and Ceratophyllaceae - which was terrestrial (Endress & Doyle 2015). Zeng et al. (2014) optimised the distribution of a large number of characters on a tree with a clade [Ceratophyllum + Chloranthaceae] (see also below). Doyle and Endress (2018) again suggest some kind of link between the two, Pseudoasterophyllites rather than Tucanopollis being involved.
2. Relationships Between the Three Major Groups, the Monocots, Magnoliids, and Eudicots.
As will have become obvious, inclusion of both Ceratophyllum and Chloranthaceae is pretty much essential in any attempt to clarify the relationships of these three major groups, which include the vast majority of angiosperms ("no CC" below means that they were not included). Thus Shen et al. (2017) evaluated the support for two hypotheses of the position of Chloranthaceae, [Magnoliids [Eudicots + Chloranthales]] vs [[Magnoliids + Chloranthaceae] Eudicots], but the situation is fluid as molecular analyses have been progressing from one to several plastome genes, to complete plastomes and transcriptomes, and now to whole nuclear genomes and transcriptomes - in large numbers of taxa. G. Zhang and Ma (2024: esp. Fig. 1) summarized the various relationships suggested by different studies up to the end of 2023 that used nuclear data. Some of all this literature is discussed below ain the context of the particular hypothesis favoured, 2A, [monocots [magnoliids + eudicots]], 2B, [eudicots [monocots + magnoliids]], 2C, [magnoliids [monocots + eudicots]], and 2D, other or all.
2A. [Monocots [magnoliids + eudicots]]. Zanis et al. (2002) analysed 11 genes from all genomes and found some support for the magnoliids (strong support for this clade) as being sister to eudicots, with Chloranthaceae, and [monocots + Ceratophyllaceae] occurring as successively more basal branches (see also Graham & Olmstead 2000; Borsch et al. 2003). Support for some of these nodes depended on the method of analysis (see also Borsch et al. 2000; Graham & Olmstead 2000; Hilu et al. 2001; Whitlock et al. 2001) or the particular gene studied (see Duvall & Bricker 2002: nuclear 18s; Hilu et al. 2003: matK; Duvall et al. 2006: nuclear 18S in particular causes problems). Soltis et al. (2007a) found very weak support for monocots as sister to [magnoliids, Chloranthaceae, eudicots, etc.]. Lee et al. (2011: Chloranthales and Ceratophyllales not included) found a clade [monocots [magnoliids + eudicots], and with strong support (see also Duarte et al. 2010), and they initially looked at almost 23,000 sets of orthologues from nuclear genomes of 101 genera of land plants (most taxa represented by ESTs, genes included if represented in as few as 4 taxa). Such relationships were strongly supported in the 59 low-copy nuclear gene analysis of Zeng et al. (2014: see also some analyses in Moore et al. 2011), and there sampling is not obviously a problem, Ceratophyllum, Sarcandra and Chloranthus being included. The [Ceratophyllum + Chloranthaceae] clade also recovered by Zeng et al. (2014) and Moore et al. (2011) was placed sister to eudicots (c.f. also Zeng et al. 2014: Table 2, p. 7 etc., conflict between single-copy and IR genes). In the nuclear and mitochondrial analyses by M. Sun et al. (2014) [Chloranthales + Ceratophyllales] were sister to monocots [check], while Morris et al. (2018) found that Chloranthaceae were sister to eudicots (Ceratophyllum not included). In an early study the relationships [Monocots [Ceratophyllum [[magnoliids + Chloranthus] eudicots]]] were obtained (Barkman et al. 2000b), although support was poor, while Zanis et al. (2002) and Y.-L. Qiu et al. (2005: other topologies also recovered) found Ceratophyllum to be sister to monocots and Chloranthus sister to the [eudicot + magnoliid] clade; relationships in Evkaikina et al. (2017) are [[[Chloranthaceae + monocots] magnoliids] eudicots]. Wickett et al. (2014) found that [magnoliids + Chloranthaceae] were sister to eudicots in coalescent transcriptome analyses, the other main topologies (2 and 3 below) being rejected. In general, this topology is more often recovered in anayses that include nuclear genes, and it was also recovered - and with quite strong support - in one of the first analyses including whole genomes of a magnoliid, Cinnamomum (Chaw et al. 2018b: no CC), but, as these authors note, improved sampling is a desideratum (see also Soltis & Soltis 2018); c.f. also the summary tree of L. Zhang et al. (2019: no CC) and Lu et al. (2020: no CC). The basic arrangement [monocots [magnoliids + eudicots]] was also recovered by O.T.P.T.I. (2019: Ceratophyllaceae usually sister to eudicots, otherwise to monocots), Y. Yang et al. (2020: no Chloranthaceae), P.-L. Liu et al. (2020: no CC), W. J. Baker et al. (2021a), Y. Liu et al. (2021: no CC), and X. Guo et al. (2021), all using nuclear genomes. This seems to be the most likely set of relationships; see also [Chloranthaceae + Magnoliids] above.
2B. [Eudicots [monocots + magnoliids]]. This topology was recovered by Hertweck et al. (2015: eight genes), with a [Ceratophyllum + Chloranthaceae] clade sister to the monocots. Eudicots were sister to an unresolved clade including monocots, magnoliids, and Chloranthaceae (D. Soltis et al. 2000; P. Soltis et al. 2000; Wu et al. 2007; Y.-L. Qiu & Estabrook 2008: compatibility analysis; Doyle & Endress 2010). Hilu et al. (2003) in a matK analysis, suggested that magnoliids were sister to monocots, although the support was weak in parsimony analyses if with 100% posterior probabilities in Bayesian analyses, furthermore, in the latter case only, [Chloranthacaeae + monocots] were sister to magnoliids, although the probabilities there were low. Davis et al. (2004) found magnoliids to be sister to monocots and that Chloranthaceae and Ceratophyllaceae were successively sister to a clade including the few eudicots in the analysis, but support was not exactly overwhelming (the best supported topology in Duvall et al. 2006 is somewhat similar). Müller et al. (2006) found very poorly supported relationships between Chloranthaceae and monocots, which together linked with the magnoliids. Whitlock et al. (2002) recovered largely similar groupings, but support was only moderate; the exact position of Chloranthaceae remained unclear. Duvall et al. (2006: four genes, three compartments) also preferred a relationship between magnoliids and monocots, as did N. Zhang et al. (2012), Doyle and Upchurch (2014) and Doyle and Endress (2014). Although Wickett et al. (2014) rejected the clade [eudicots [monocots + magnoliids]] in their transcriptome study, it was recovered by Zhao et al. (2020, 2021) in their microsynteny analyses. L. Qin et al. (2021) also found that topologies recovered in sequence-based analyses depended on the analyses, again there being rapid divergence involving magnoliids, monocots, etc., ca 200 Ma, however, Qin et al. noted structural changes in the genome of Aristolochia fimbriata that were shared both with other magnoliids and with monocots. Interestingly, Qin et al. (2021) found no evidence of genome duplication events involving angiosperms like A. fimbriata (their study system), Nymphaea and Amborella, which is of course of some general interest.
2C. [Magnoliids [monocots + eudicots]]. A [monocot + eudicot] grouping has quite often been recovered, as in Jansen et al. (2006b: 37 whole chloroplast genomes, see also Zhengqiu et al. 2006), Cai et al. (2006: 35 whole chloroplast genomes, no CC or Austrobaileyales), Duvall et al. (2006: four genes, three compartments, nuclear PHYC gene, 18S being excluded), Mathews (2006a: three PHY genes, 105 taxa), Hansen et al. (2007: 61 protein-coding chloroplast genes), Mardanov et al. (2008: ?sampling), Goloboff et al. (2009), Xue et al. 2012: whole chloroplast genomes), Z.-D. Chen et al. (2016), Givnish et al. (2016b: 75 plastid genes: stem monocots (146-)142(-136) My) , Foster et al. (2017: support weak), Zhong and Betancur R. (2017: no CC), Gitzendanner et al. (2018a: plastid genomes, support weak); Rendón-Anaya et al. (2019: no CC, nuclear genomes, the predominant topology) and Ramírez-Barahona et al. (2020: three nuclear ribsomnal and four chloroplast genes). Graham et al. (2005) found a rather weakly supported (73% bootstrap) [monocot + eudicot] grouping, and this was yet more weakly supported when Chloranthaceae and Ceratophyllaceae were included. When sequences of complete chloroplast genomes are analysed, an association between monocots and eudicots may be more strongly suggested. Thus Jansen et al. (2007) found good support for a [monocot + eudicot] grouping, a number of alternative topologies being excluded, but support in Moore et al. (2007) and Foster et al. (2016) was somewhat less strong; see also Magallón et al. (2015). Although the possibility of a [monocot + eudicot] grouping was rejected by Wickett et al. (2014: transcriptome analyses), this was the preferred topology in the anchored phylogenomic analysis of Buddenhagen et al. (2016: no CC), especially in coalescent-based analyses (there was some support for a [eudicot + magnoliid] clade in supermatrix analyses, but only when all data were included, the support disappearing when rapidly-evolving sites were removed). H.-T. Li et al. (2019, see also 2021) recovered these relationship in their chloroplast genome analyses, although support for the [monocot [Ceratophyllum + eudicot]] clade was only 72% bootstrap, and also in the plastome analyses of Y. Yang et al. (2020). Interestingly, this basic topology was also recovered in two recent analyses including whole genomes of magnoliids, Liriodendron (J. Chen et al. 2018: support rather weak) and Piper (L. Hu et al. 2019: no CC, support?), but improved sampling is essential (see also Soltis & Soltis 2019). Indeed, some other genome analyses with better, if still sometimes somewhat exiguous sampling, have found the relationships [monocots [magnoliids + eudicots]], see 2A above.
Evolution of some floral developmental genes, e.g. in the C and D lineages, are also consistent with a [monocot + eudicot] clade (Kramer et al. 2004), as is the distribution of taxa in which DEF-like proteins cannot form heterodimers (Melzer et al. 2014) and of taxa with a common regulatory network governing floral symmetry (Madrigal et al. 2019:: not the DIVARICATA transcription factor analysed singly). Variation in how calcium oxalate is synthesized may also be of phylogenetic interest, although sampling is very poor; neither members of the ANITA grade nor magnoliids have been examined. Vacuolar crystal formation associated with membranes and paracrystalline bodies with widely spaced subunits are found in eudicots (crystals seem also to be formed in other ways here), while in monocots there are no membrane complexes and the paracrystalline bodies have closely spaced subunits (Horner & Wagner 1995; Evert 2006). Some patterns of RNA editing may be phylogenetically informative at this level (Logacheva et al. 2008). The positional relationships between members of the androecium and the perianth, the stamens being individually opposite perianth members, and also trimery of some floral whorls, occur in a number of taxa, and their distribution is broadly consistent with all three sets of relationships.
2D. Other, or All, Relationships. Analysis of morphological data in particular has quite often grouped more or less herbaceous clades in magnoliids and the ANITA grade, i.e. Piperales, Chloranthaceae, and Nymphaeales, with monocots; this is the pal(a)eoherb hypothesis (see e.g. Donoghue & Doyle 1989; Taylor & Hickey 1992; Endress 2000). Analysis of combined morphological and molecular data suggested that Piperales were sister to monocots (Doyle & Endress 2000: support weak), Tamura et al. (2011: 6 plastid, 7 mitochondrial, and 1 nuclear genes sequenced) found some support for this clade, as did Barrett and Davis (2011: or [Piperales + Ceratophyllales] the sister group). However, although quite popular in the latter years of the last century, the palaeoherb support comes from homoplasies associated with the adoption of the herbaceous habit. Nymphaeaceae s.l., i.e. including Nelumbonaceae and Cabombaceae, have more or less dorsiventral embryos, in this rather like the embryos of Ceratophyllum and monocots (see Titova & Batygina 1996). Nevertheless, Piperales are now firmly (usually!) embedded in the magnoliid clade, suggestions like that of Parkinson et al. (1999) of a [Piperales + eudicot] and by Ruhfel et al. (2014: amino acid analyses) of a [Piperales + Ceratophyllaceae] clade being exceptions, while Nymphaeales have found a resting place very near the base of the whole angiosperm clade.
J. Chen et al. (2018) looked at the genomes of Liriodendron and sixteen other angiosperms and found almost identical numbers of orthogroups, i.e. groups of genes descended from a gene in the common ancestor of these species, and this supported all three hypotheses above... Rendón-Anaya et al. (2019) focussed on the genome of Persea and also obtained all three topologies, depending on whether coding or protein sequences were analysed, etc., and they suggested that one problem was the short time, perhaps just 4 Ma, between the separation of the three main groups. Y.-C. Chen et al. (2020: no CC) did not particularly prefer any of the three hypotheses above in the various analyses that they carried out - again, related factors like time, ILS, etc., were issues.
To Conclude. Shen et al. (2023: analytical categories unclear) summarised recent work in this area, [magnoliids [monocots + eudicots]] and [magnoliids + eudicots] were the favoured hypotheses. From the survey here, it seems that there may be be differing signals in nuclear (relationships - [monocots [magnoliids + eudicots]]) and chloroplast ([magnoliids + [monocots + eudicots]]) genes (Xi et al. 2014: no CC; see also X. Guo et al. 2021, etc.). Sun et al. (2014) retrieved the latter set of relationships, but analyses of both nuclear and mitochondrial trees separately recovered a [eudicots [monocots + magnoliids]] topology. Wickett et al. (2014) in particular noted that analyses of chloroplast genes did not give consistent relationships. MP and ML analyses may also give different topologies (Bauscher et al. 2006). Of course, Ceratophyllum and Chloranthales must be included in future analyses of the relationships between the monocots, magnoliids and eudicots; in their absence, the analyses must be considered incomplete. Overall, basal mesangiosperm relationships may well be [monocots [[Chloranthaceae + magnoliids] [Ceratophyllum + eudicots]]], and that is the topology followed in the text here.
Indeed, comparing the relationships suggested by three groups for taxa in this area is interesting. 1. APG IV (2016: largely chloroplast data): [Chloranthales, [[Canellales + Piperales] [Magnoliales + Laurales]], [Monocots [Ceratophyllales [Eudicots ... (H.-T. Li et al. 2021 - plastome data - is similar, except that the tritomy is no more). 2. Zuntini et al. (2024: Angiosperms353 nuclear data): ... [Ceratophyllales [Monocots [Chloranthales [[Canellales + Piperales]] [Magnoliales + Laurales]]] [Eudicots... Support for the position of Ceratophyllales is very weak. 3. Y.-L. Qiu et al. (2024: 1 nuclear, 2 plastid, 2 mitochondrial markers): ...[Monocots [[Ceratophyllales + Chloranthales] [[[Canellales + Piperales] [Magnoliales + Laurales]] [Eudicots...[[CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]: benzylisoquinoline alkaloids +.
Age. This node is ca 159.6 Ma (X. Guo et al. 2021).
[CHLORANTHALES [[MAGNOLIALES + LAURALES] [CANELLALES + PIPERALES]]]: sesquiterpenes +; (microsporogenesis also simultaneous); seed endotestal.
Age. Moore et al. (2010) estimated an age of (141-)136(-129) Ma for this node and Xue et al. (2012) an age of ca 143.2 or 141.3 Ma. Clarke et al. (2011: other estimates) a somewhat older age of (176-)149(-128) Ma, Soltis et al. (2008) suggested (168-)131(-126) Ma, and Magallón et al. (2013) an age of around 149.1 Ma and Naumann et al. (2013) an age of around 142.5 Ma; see also Magallón (2009) for other dates around 140-150 Ma, about 134.6 Ma was estimated by Magallón et al. (2015), ca 135.5 Ma by X. Guo et al. (2021), ca 170 Ma by Foster et al. (2016, q.v. for details), ca 198.9 to 192 Ma by Tank et al. (2015: Table S1, S2) and (221-)194(-169) Ma by Salomo et al. (2017: also other dates). The estimate in Z. Wu et al. (2014), at ca 210 Ma, is the oldest.
Evolution: Divergence & Distribution. Soltis et al. (2008) offer ages for a number of branching points within this clade based on the topology [monocots [Chloranthaceae, magnoliids [Ceratophyllaceae + eudicots]]].
The character, "endotesta palisade, crystaliferous", could perhaps be placed at this node. The radially elongated endotestal cells reflect the presence of an endothelium in the ovule, and Friis et al. (2019b) think that this may have been common in early angiosperms.
Chemistry, Morphology, etc.. Microsporogenesis is variable throughout this clade (Furness & Rudall 2004).
CHLORANTHALES Martius - Main Tree.
Just the one family, 4 genera, 75 species.
Note: In all node characterizations, boldface denotes a possible apomorphy, (....) denotes a feature the exact status of which in the clade is uncertain, [....] includes explanatory material; other text lists features found pretty much throughout the clade. However, the precise node to which many characters, particularly the more cryptic ones, should be assigned is unclear. This is partly because homoplasy is very common, in addition, basic information for all too many characters is very incomplete, frequently coming from taxa well embedded in the clade of interest and so making the position of any putative apomorphy uncertain. Then there are the not-so-trivial issues of how character states are delimited and ancestral states are reconstructed (see above).
Includes Chloranthaceae.
Synonymy: Chloranthineae Thorne & Reveal - Chloranthanae Doweld - Chloranthidae C. Y. Wu
CHLORANTHACEAE Sims, nom. cons. - Back to Chloranthales
Evergreen shrubs or trees; neolignans ?+; primary stem with vascular cylinder/not; rays 6-10-seriate; elongated mucilage ducts +; (sclereids - Hedyosmum); cuticle wax crystalloids 0; branching from current flush, nodes often swollen; leaves opposite, joined by sheath, lamina vernation conduplicate [Chloranthus], margins serrate, teeth with clear persistent swollen cap into which proceed higher order veins as well as secondaries or tertiaries [hydathodal]; stipules small, paired, interpetiolar, usually on rim of sheath; inflorescence spicate (branched), flowers sessile, very small [4> mm across], monosymmetric by reduction, parts whorled; P 0; staminate flowers: pistillode 0; carpelate flowers: G 1, ± inferior, ascidiate, postgenital fusion by secretion, [compitum necessarily 0], style 0, stigma with ad-/abaxial lobes, dry; ovule 1/carpel, apical, pendulous, straight, outer integument 4-8 cells across, inner integument (3-)7-10 cells across, endothelium +, (micropyle bistomal), parietal tissue 6-8 cells across, nucellar cap +/0; antipodal cells proliferating; fruit baccate or drupaceous, (bracts accrescent and succulent), (P persistent); coat ± tanniniferous, endotesta ± palisade, lignified, containing crystals, with endoreticulate fibres, tegmen ± crushed, exotegmen fibrous, endotegmen initially subpalisade; endosperm cellular, starchy, grains clustered); x = 7 (?8), chromosomes 1-4(-10) µm long, nuclear genome [1 C] (0.029-)2.368(-196.237) pg.
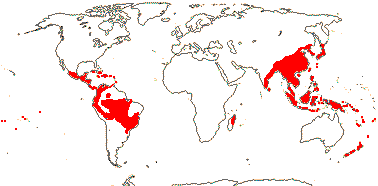
4 [list]/75: Hedyosmum (45). Tropics and subtropics, not Africa (Madagascar - Ascarina only) (map: from Verdcourt 1986; Todzia 1988; Leitman 2014). [Photo - Leaf, Flower.]
Age. Magallón and Castillo (2009) estimated the crown group age of Chloranthaceae to be ca 153.6 or 125 Ma, Wikström et al. (2001) date it at 131-121 Ma, Bell et al. (2010) at around 121 or around 98 Ma, depending on the analysis, Antonelli and Sanmartín (2011: fossil-based) suggested ages of (112-)111(-110) Ma or thereabouts, Salomo et al. (2017) an age of (125-)117(-113) Ma, one of several estimates, while Zhang et al. (2011) provided yet another series of different age estimates, some of which are dramatically older than the others depending on the calibration and the analytical methods used.
Fossils assignable to Chloranthaceae were already common, diverse, and world-wide in distribution when angiosperms first appeared in the fossil record (Friis et al. 2011, 2015a, 2019c, esp. 2019f and 2022b and references, 2022, 2024; Doyle & Upchurch 2014; Tekleva et al. 2021). Distinctive fossil pollen grains, Asteropollis, are first known from the Barremian-Aptian of the early Cretaceous, some 125 Ma (Friis et al. 1997; Doyle 1999; Eklund 1999, but c.f. Clarke et al. 2011, questions over dating); these grains have been identified as Hedyosmum (see also Crepet & Nixon 1996; Eklund et al. 2003; Friis et al. 2005; Martínez et al. 2013; Doyle & Endress (2018), c.f. age estimates in preceding paragraph). Doyle and Endress (2007), Clarke et al. (2011) and Friis et al. (2024) discuss other palynomorphs that have been associated with Chloranthaceae including Clavatipollenites. Some fossil androecia assigned to the family have spiraperturate pollen that has been found in situ (Crane et al. 1989); described as Chloranthistemon endressii, the identity of this fossil as a member of Chloranthaceae, either in Chloranthus (Doyle & Endress 2018) or as closer to Sarcandra (Schönenberger et al. 2020) has been confirmed, despite the fact that the fossil is very incomplete (see also López-Martínez et al. 2023a: Fig. 2A), also, there is the Late Barremian to Aptian Canrightia resinifera to think about (ibid., but see also Thismiaceae and Stemonaceae). Similarly, the pollen morph Clavatipollenites, widespread in the fossil record in the Cretaceous, is like that of Ascarina but it has also been found on fruits of Couperites that may not belong to the family (Doyle et al. 2003). Pollen of the Catefica fossil (= ) is described and discussed in detail by Tekleva et al. (2021); this fossil was generally associated with Hedyosmum, although in some analyses it was also placed elsewhere in Chloranthaceae or with Ceratophyllum, whether that genus was also associated with Chloranthaceae or sister to eudicots, and it might jump to Nymphaeales in trees one or two steps longer. Flowers, fruits and seeds of Chloranthaceae are also known fossil, especially from deposits in Portugal, but also from North America - see Friis et al. (2018b) for literature and also descriptions of two more genera of chloranthoid seeds. Pollen of Asteropollis has been found associated with Hedyflora, but Friis et al. (2019c) suggested that it may eventually be found to be associated with several different kinds of flowers. Doyle and Endress (2010) thought that the Pennipollis plant, from ca 120-115 Ma in Portugal and originally associated with the monocots (Petersen et al. 2000b), was sister to Chloranthaceae. See Friis et al. (2024) for a detailed evaluation of the fossil record.
Tracheids ca 940 µm long; nodes 1:4; stomata stephanocytic; plant dioecious (monoecious), flowers often rather densely aggregated; staminate flower: bract 0; P +, uniseriate, ± connate; anther connective truncate to triangular to massive and 3-lobed, hypodermis lignified; pollen with branched sulcus [4-6-branched], supratectal spinules +; pistillode 0; carpellate flowers: bract +, large, fleshy; ?P 3-lobed, connate; staminodes 0; style +, stigma caducous; endothelium 0; extensive hypanthial outgrowth, aril-like, carpel wall largely 0; endotesta undifferentiated, exotegmic fibres 0; n = 8, chromosomes to 10 µm long.
1/41. Largely Tropical America, inc. the Caribbean, Hedyosmum orientale S. China, Hainan, South Vietnam, Indonesia.
[Ascarina [Chloranthus + Sarcandra]]: stomata with outer rims raised; P 0; hypanthium reduced; carpel wall lacking crystals; inner integument 3 or more cells across.
Age. The age of this clade is around 46.0 Ma (X. Guo et al. 2021).
2. Ascarina J. R. & G. Forster
Tracheids ca 940 µm long; nodes 1:2; stomata encyclocytic; plant dioecious (monoecious); staminate flowers: bract +; A 2, or bract +, bracteoles +; A 1-5; connective inconspicuous, hypodermis lignified; pollen monosulcate, supratectal spinules +; pistillode +; outer integument ca 2 cells across; exotegem fibrous to sclerotic, endotegmen tanniniferous [?= endothelium]; n = 14.
1/10. Madagascar, Malesia and New Zealand to the Pacific.
[Chloranthus + Sarcandra]: plants subshrubs; nodes with split laterals; flowers perfect; bract +; A abaxial, adnate to G, anthers dehiscing by lateral valves; stigma symmetrical; carpel wall with intrusive oil cells; endothelium 0; exotegmen fibres 0; n = 15.
Age. This clade is ca 32.2 Ma (X. Guo et al. 2021).
(Rhizomatous herbs); tracheids ca 940 µm long; stomata paracytic (anomocytic); (leaves pseudoverticillate); A trilobed, adnate to dorsal G, bases connate, connective lobes ± conspicuous (much elongated), thecae reduced, median lobe 2-thecate, 4-locular, lateral lobes 1-thecate, 2-locular; ?vascular supply]; pollen 3-8-colpate/pantoporate (trichotomosulcate); stigma small; mesotesta lignified; n = (14).
1/17. Temperate and tropical Asia, Malesia.
Vessels only in root secondary xylem and stem metaxylem; tracheids ca 2000 µm long; reaction wood on lower side of stem; nodes 1:4; stomata anomocytic; A 1, anther connective short-truncate, filament relatively long, wide; pollen pantoporate.
1/2. China and India to Malesia.
Evolution: Divergence & Distribution. Pollen compared with that of Ascarina has been reported from southwest South African deposits only some 15 Ma (Coetzee & Muller 1984); there are of course currently no Chloranthaceae in Africa and environs.
With older estimates of the family age, all genera had separated by ca 90 Ma (e.g. Wikström et al. 2000; Antonelli & Sanmartín 2011). However, other suggestions are that crown group diversification of the genera is relatively recent, being mostly within the last 60 Ma (Zhang & Renner 2003b; Soltis et al. 2008; Antonelli & Sanmartín 2011). Thus the evolutionary stems of the genera are very long, for instance, some 60-83.5 Ma when using the estimates of Antonelli and Sanmartín (2011); they thought that diversification within Hedyosmum could be dated to (57.1-)43.3(-30.1) or (43-)35.6-(25.9) Ma depending on the method used, and was in part associated with the Andean uplift. However, the family as a whole showed a pattern of gradual extinction over time.
For general morphological evolution, living and fossil Chloranthaceae integrated, see Eklund et al. (2004) and especially Friis et al. (2024). Endress (2001) emphasized what he considered to be the plesiomorphic floral morphology of the family. However, there is no evidence that it is a member of the ANA grade, and several aspects of its flowers and their development, including the loss of any perianth, are clearly derived (e.g. G.-S. Li et al. 2005). Indeed, Sarcandra chloranthoides is one of the five angiosperms with the greatest floral eccentricities, i.e. divergence from the average, examined by López-Martínez et al. (2023b). Friis et al. (2019b) discuss the possibility that an endothelium is plesiomorphic for the family, noting that fossils like Canrightia and Rightcania have a crystaliferous endotesta/endothelium, the cells being more or less radially elongated and crystaliferous (Friis et al. 2018b). For the evolution of the chloranthaceous flower, integrating fossils into the scenario, see Doyle and Endress (2014). It has been suggested that the flowers of Chloranthus and Sarcandra are secondarily perfect/bisexual (see also Doyle & Endress 2011), or imperfect flowers have evolved twice, or perfect flowers are really pseudanthia (Endress & Doyle 2015)... In the careful analysis of Friis et al. (2015a), Canrightiopsis, with a single carpel and three stamens borne on one side of semi-inferior gynoecium, shows similarities with Canrightia; Doyle and Endress (2018) recovered the relationships [Canrightiopsis [Sarcandra + Chloranthus]]. Canrightia itself has a hypanthium, semi-inferior ovary, 2-5 carpels each with single ovules, the ovules perhaps with an endothelium (the endotegmic cells are radially elongated), a crystal-bearing endotesta, and its stamens are radially arranged; at 126-110 Ma, it attaches to the stem of Chloranthaceae in phylogenetic analyses (Friis & Pedersen 2011). The seeds of Canrightiopsis are very small, and its embryo is about one quarter the size of that of Sarcandra, with which it has been compared (Friis et al. 2015b).
Ecology & Physiology. For possible ecophysiological and morphological changes here associated with the κ duplication, and in more or less contemporaneous duplications in Magnoliales + Laurales, Nymphaeales, the core eudicots, and in the monocots, perhaps at the [Asparagales + commelinds] node, see the elsewhere.
Sarcandra glabra lacks vessels and has - unusually for angiosperms - compression wood, and this may be associated with the fact that the ratio of syringyl to guaiacyl units in the lignin is low, as in Pinales, which also have compression wood (Aiso et al. 2014, 2016).
Pollination Biology & Seed Dispersal. Nectar is reported from Sarcandra at least, and pollination may be by small insects (and in Chloranthus), while wind pollination occurs in Ascarina and Hedyosmum (Erbar 2014; Gottsberger 2016a and references). Stigmatic self incompatability reactions have been recorded from Chloranthaceae (Allen & Hiscock 2008).
Plant-Animal Interactions. There has beeen quite a substantial radiation of the geometrid moth Eios (the Adimaria clade) on
Genes & Genomes. There may have been a genome duplication event at the [Sarcandra + Ascarina] node some 106.9 Ma (Landis et al. 2018), while X. Guo et al. (2021) date the κ/kappa duplication event in the family at 130.5-98.1 Ma - this duplication may be involved in the evolution of stress-resistance genes.
Chemistry, Morphology, etc.. Although benzylisoquinoline alkaloids apparently have not been detected in Chloranthaceae, (S)norcolaurine synthase activity is high, suggesting that they may be found here (Liscombe et al. 2005).
There are eight vascular bundles in younger stems, four from the node immediately above and four from the node above that; the stele is basically foliar (Balfour & Philipson 1962). There are reports of endodermis in the stem here (Seago 2020). Roots - presumably those of the seedlings and young plants - seem not have any secondary thickening (Blanc 1986)?
There is some discussion of inflorescence morphology in Doria et al. (2012). Tekleva et al. (2021) discuss a number of aspects of pollen and flower. The perianth of Hedyosmum has unique, schizogenous apertures/windows (Doria et al. 2012 and references; c.f. Endress & Doyle 2015). Swamy (1953a), Crane et al. (1989), Crepet and Nixon (1997), Eklund et al. (1997), Eklund (1999), and others discuss the nature of the androecium in the family, a matter over which there has been considerable controversy. In Chloranthus it has been suggested that the androecium has 2 or 4 dithecal stamens or that it consists of a single 8-locular stamen or..., and that staminate flowers of Hedyosmum have hundreds of anthers. Here, as in most other angiosperms, the stamens seem to be dithecal (Swamy 1953a; Kong et al. 2002, and references). Gabarayeva et al. (2021) discussed the development of the pollen wall - largely by self-assembly - of Chloranthus japonicus. Doria et al. (2012) illustrate a superior gynoecium in Hedyosmum. The stylulus is filled with secretion. Endress and Igersheim (1997) describe the stigma as being wet (c.f. Todzia 1988). The ovule in Chloranthus is not quite straight but "subatropous", according to Yamada et al. (2001a). For variation in micropyle type, see Heo and Tobe (1995). Friis et al. (2019c) described the exotesta of the fossil Hedyflora, in several respects close to Hedyosmum, as being made up of longitudinal fibres. Johri et al. (1992) noted that the endosperm stores oil, but there may also be starch. The cotyledonary nodes have split laterals (Bailey 1956).
For general information, see Swamy (1953a: comparison with 18 families, only one close, 1953c: Ascarina), Todzia (1988, 1993) and Eklund (1999), for chemistry, see Hegnauer (1964, 1989), young plants, see Blanc (1986), general vegetative anatomy, see Datta and Dasgupta (1977a, b) and Metcalfe (1987), for xylem/wood anatomy, see Carlquist (1987c: Sarcandra, 1992a), for embryology, see Vijayaraghavan (1964), and for floral development, see Endress (1987b).
Phylogeny. Within Chloranthaceae, morphological analyses, including details of wood anatomy, suggested the genus pairs [Ascarina + Hedyosmum], mainly woody, plant monoecious to dioecious, and [Chloranthus + Sarcandra], herbaceous to semi-shrubby, flowers perfect. In early molecular work (Qiu et al. 1999), on the other hand, the relationships [Hedyosmum [Ascarina [Chloranthus + Sarcandra]]] were found. Although there was strong support, the sampling was rather minimal, but the same set of relationships were confirmed by Zhang and Renner (2003b) and Zhang et al. (2011), both with a much improved sampling. They have also been found in subsequent morphological analyses with constrained outgroups (Doyle et al. 2003; Eklund et al. 2004: see esp. Figs 10, also 11-15, and Table 3). However, in the admittedly preliminary Angiosperm353 analysis relationships were different yet again, being [Chloranthus [Hedyosmum [Ascarina + Sarcandra] (W. J. Baker et al. 2021a: see Seed Plant Tree of Life, four species sampled).
Previous Relationships. Cronquist (1981) included Chloranthaceae in his Piperales while Takhtajan (1997) placed Chloranthales after Calycanthales at the end of his Magnoliidae.
Synonymy: Hedyosmaceae Caruel