EMBRYOPSIDA Pirani & Prado
Gametophyte dominant, independent, multicellular, initially ± globular, not motile, branched; showing gravitropism; glycolate oxidase +, glycolate metabolism in leaf peroxisomes [glyoxysomes], acquisition of phenylalanine lysase* [PAL], flavonoid synthesis*, microbial terpene synthase-like genes +, triterpenoids produced by CYP716 enzymes, CYP73 and phenylpropanoid metabolism [development of phenolic network], xyloglucans in primary cell wall, side chains charged; plant poikilohydrous [protoplasm dessication tolerant], ectohydrous [free water outside plant physiologically important]; thalloid, leafy, with single-celled apical meristem, tissues little differentiated, rhizoids +, unicellular; chloroplasts several per cell, pyrenoids 0; centrioles/centrosomes in vegetative cells 0, microtubules with γ-tubulin along their lengths [?here], interphase microtubules form hoop-like system; metaphase spindle anastral, predictive preprophase band + [with microtubules and F-actin; where new cell wall will form], phragmoplast + [cell wall deposition centrifugal, from around the anaphase spindle], plasmodesmata +; antheridia and archegonia +, jacketed*, surficial; blepharoplast +, centrioles develop de novo, bicentriole pair coaxial, separate at midpoint, centrioles rotate, associated with basal bodies of cilia, multilayered structure + [4 layers: L1, L4, tubules; L2, L3, short vertical lamellae] (0), spline + [tubules from L1 encircling spermatid], basal body 200-250 nm long, associated with amorphous electron-dense material, microtubules in basal end lacking symmetry, stellate array of filaments in transition zone extended, axonemal cap 0 [microtubules disorganized at apex of cilium]; male gametes [spermatozoids] with a left-handed coil, cilia 2, lateral, asymmetrical; oogamy; sporophyte +*, multicellular, growth 3-dimensional*, cuticle +*, plane of first cell division transverse [with respect to long axis of archegonium/embryo sac], sporangium and upper part of seta developing from epibasal cell [towards the archegonial neck, exoscopic], with at least transient apical cell [?level], initially surrounded by and dependent on gametophyte, placental transfer cells +, in both sporophyte and gametophyte, wall ingrowths develop early; suspensor/foot +, cells at foot tip somewhat haustorial; sporangium +, single, terminal, dehiscence longitudinal; meiosis sporic, monoplastidic, MTOC [= MicroTubule Organizing Centre] associated with plastid, sporocytes 4-lobed, cytokinesis simultaneous, preceding nuclear division, quadripolar microtubule system +; wall development both centripetal and centrifugal, 1000 spores/sporangium, sporopollenin in the spore wall* laid down in association with trilamellar layers [white-line centred lamellae; tripartite lamellae]; plastid transmission maternal; nuclear genome [1C] <1.4 pg, main telomere sequence motif TTTAGGG, KNOX1 and KNOX2 [duplication] and LEAFY genes present, ethylene involved in cell elongation; chloroplast genome with close association between trnLUAA and trnFGAA genes [precursors for starch synthesis], tufA, minD, minE genes moved to nucleus; mitochondrial trnS(gcu) and trnN(guu) genes +.
Many of the bolded characters in the characterization above are apomorphies of more or less inclusive clades of streptophytes along the lineage leading to the embryophytes, not apomorphies of crown-group embryophytes per se.
All groups below are crown groups, nearly all are extant. Characters mentioned are those of the immediate common ancestor of the group, [] contains explanatory material, () features common in clade, exact status unclear.
POLYSPORANGIOPHYTA†
Sporophyte well developed, branched, branching dichotomous, potentially indeterminate; hydroids +; stomata on stem; sporangia several, terminal; spore walls not multilamellate [?here].
II. TRACHEOPHYTA / VASCULAR PLANTS
Sporophyte long lived, cells polyplastidic, photosynthetic red light response, stomata open in response to blue light; plant homoiohydrous [water content of protoplasm relatively stable]; control of leaf hydration passive; plant endohydrous [physiologically important free water inside plant]; PIN[auxin efflux facilitators]-mediated polar auxin transport; (condensed or nonhydrolyzable tannins/proanthocyanidins +); borate cross-linked rhamnogalactan II, xyloglucans with side chains uncharged [?level], in secondary walls of vascular and mechanical tissue; lignins +; roots +, often ≤1 mm across, root hairs and root cap +; stem apex multicellular [several apical initials, no tunica], with cytohistochemical zonation, plasmodesmata formation based on cell lineage; vascular development acropetal, tracheids +, in both protoxylem and metaxylem, G- and S-types; sieve cells + [nucleus degenerating]; endodermis +; stomata numerous, involved in gas exchange; leaves +, vascularized, spirally arranged, blades with mean venation density ca 1.8 mm/mm2 [to 5 mm/mm2], all epidermal cells with chloroplasts; sporangia in strobili, sporangia adaxial, columella 0; tapetum glandular; sporophyte-gametophyte junction lacking dead gametophytic cells, mucilage, ?position of transfer cells; MTOCs not associated with plastids, basal body 350-550 nm long, stellate array in transition region initially joining microtubule triplets; archegonia embedded/sunken [only neck protruding]; embryo suspensor +, shoot apex developing away from micropyle/archegonial neck [from hypobasal cell, endoscopic], root lateral with respect to the longitudinal axis of the embryo [plant homorhizic].
[MONILOPHYTA + LIGNOPHYTA]Sporophyte growth ± monopodial, branching spiral; roots endomycorrhizal [with Glomeromycota], lateral roots +, endogenous; G-type tracheids +, with scalariform-bordered pits; leaves with apical/marginal growth, venation development basipetal, growth determinate; sporangium dehiscence by a single longitudinal slit; cells polyplastidic, MTOCs diffuse, perinuclear, migratory; blepharoplasts +, paired, with electron-dense material, centrioles on periphery, male gametes multiciliate; nuclear genome [1C] 7.6-10 pg [mode]; chloroplast long single copy ca 30kb inversion [from psbM to ycf2]; mitochondrion with loss of 4 genes, absence of numerous group II introns; LITTLE ZIPPER proteins.
LIGNOPHYTA†
Sporophyte woody; stem branching axillary, buds exogenous; lateral root origin from the pericycle; cork cambium + [producing cork abaxially], vascular cambium bifacial [producing phloem abaxially and xylem adaxially].
SEED PLANTS† / SPERMATOPHYTA†
Growth of plant bipolar [plumule/stem and radicle/root independent, roots positively geotropic]; plants heterosporous; megasporangium surrounded by cupule [i.e. = unitegmic ovule, cupule = integument]; pollen lands on ovule; megaspore germination endosporic, female gametophyte initially retained on the plant, free-nuclear/syncytial to start with, walls then coming to surround the individual nuclei, process proceeding centripetally.
EXTANT SEED PLANTS
Plant evergreen; nicotinic acid metabolised to trigonelline, (cyanogenesis via tyrosine pathway); microbial terpene synthase-like genes 0; primary cell walls rich in xyloglucans and/or glucomannans, 25-30% pectin [Type I walls]; lignin chains started by monolignol dimerization [resinols common], particularly with guaiacyl and p-hydroxyphenyl [G + H] units [sinapyl units uncommon, no Maüle reaction]; roots often ≥1 mm across, stele diarch to pentarch, xylem and phloem originating on alternating radii, cork cambium deep seated, gravitropism response fast; stem apical meristem complex [with quiescent centre, etc.], plasmodesma density in SAM 1.6-6.2[mean]/μm2 [interface-specific plasmodesmatal network]; eustele +, protoxylem endarch, endodermis 0; wood homoxylous, tracheids and rays alone, tracheid/tracheid pits circular, bordered; mature sieve tube/cell lacking functioning nucleus, sieve tube plastids with starch grains; phloem fibres +; cork cambium superficial; leaf nodes 1:1, a single trace leaving the vascular sympodium; leaf vascular bundles amphicribral; guard cells the only epidermal cells with chloroplasts, stomatal pore with active opening in response to leaf hydration, control by abscisic acid, metabolic regulation of water use efficiency, etc.; branching by axillary buds, exogenous; prophylls two, lateral; leaves with petiole and lamina, development basipetal, lamina simple; sporangia borne on sporophylls; spores not dormant; microsporophylls aggregated in indeterminate cones/strobili; grains monosulcate, aperture in ana- position [distal], primexine + [involved in exine pattern formation with deposition of sporopollenin from tapetum there], exine and intine homogeneous, exine alveolar/honeycomb; ovules with parietal tissue [= crassinucellate], megaspore tetrad linear, functional megaspore single, chalazal, sporopollenin 0; gametophyte ± wholly dependent on sporophyte, development initially endosporic [apical cell 0, rhizoids 0, etc.]; male gametophyte with tube developing from distal end of grain, male gametes two, developing after pollination, with cell walls; embryo cellular ab initio, suspensor short-minute, embryonic axis straight [shoot and root at opposite ends], primary root/radicle produces taproot [= allorhizic], cotyledons 2; embryo ± dormant; chloroplast ycf2 gene in inverted repeat, trans splicing of five mitochondrial group II introns, rpl6 gene absent; ??whole nuclear genome duplication [ζ/zeta duplication event], 2C genome size (0.71-)1.99(-5.49) pg, two copies of LEAFY gene, PHY gene duplications [three - [BP [A/N + C/O]] - copies], 5.8S and 5S rDNA in separate clusters.
IID. ANGIOSPERMAE / MAGNOLIOPHYTA
Lignans, O-methyl flavonols, dihydroflavonols, triterpenoid oleanane, apigenin and/or luteolin scattered, [cyanogenesis in ANA grade?], lignin also with syringyl units common [G + S lignin, positive Maüle reaction - syringyl:guaiacyl ratio more than 2-2.5:1], hemicelluloses as xyloglucans; root cap meristem closed (open); pith relatively inconspicuous, lateral roots initiated immediately to the side of [when diarch] or opposite xylem poles; epidermis probably originating from inner layer of root cap, trichoblasts [differentiated root hair-forming cells] 0, hypodermis suberised and with Casparian strip [= exodermis]; shoot apex with tunica-corpus construction, tunica 2-layered; starch grains simple; primary cell wall mostly with pectic polysaccharides, poor in mannans; tracheid:tracheid [end wall] plates with scalariform pitting, multiseriate rays +, wood parenchyma +; sieve tubes enucleate, sieve plates with pores (0.1-)0.5-10< µm across, cytoplasm with P-proteins, not occluding pores of plate, companion cell and sieve tube from same mother cell; ?phloem loading/sugar transport; nodes 1:?; dark reversal Pfr → Pr; protoplasm dessication tolerant [plant poikilohydric]; stomata randomly oriented, brachyparacytic [ends of subsidiary cells ± level with ends of guard cells], outer stomatal ledges producing vestibule, reduction in stomatal conductance with increasing CO2 concentration; lamina formed from the primordial leaf apex, margins toothed, development of venation acropetal, overall growth ± diffuse, secondary veins pinnate, fine venation hierarchical-reticulate, (1.7-)4.1(-5.7) mm/mm2, vein endings free; flowers perfect, pedicellate, ± haplomorphic, protogynous; parts free, numbers variable, development centripetal; P = T, petal-like, each with a single trace, outer members not sharply differentiated from the others, not enclosing the floral bud; A many, filament not sharply distinguished from anther, stout, broad, with a single trace, anther introrse, tetrasporangiate, sporangia in two groups of two [dithecal], each theca dehiscing longitudinally by a common slit, ± embedded in the filament, walls with at least outer secondary parietal cells dividing, endothecium +, cells elongated at right angles to long axis of anther; tapetal cells binucleate; microspore mother cells in a block, microsporogenesis successive, walls developing by centripetal furrowing; pollen subspherical, tectum continuous or microperforate, ektexine columellate, endexine restricted to the apertural regions, thin, compact, intine in apertural areas thick, orbicules +, pollenkitt +; nectary 0; carpels present, superior, free, several, spiral, ascidiate [postgenital occlusion by secretion], stylulus at most short [shorter than ovary], hollow, cavity not lined by distinct epidermal layer, stigma ± decurrent, carinal, dry; suprastylar extragynoecial compitum +; ovules few [?1]/carpel, marginal, anatropous, bitegmic, micropyle endostomal, outer integument 2-3 cells across, often largely subdermal in origin, inner integument 2-3 cells across, often dermal in origin, parietal tissue 1-3 cells across, nucellar cap?; megasporocyte single, hypodermal, functional megaspore lacking cuticle; female gametophyte lacking chlorophyll, four-celled [one module, egg and polar nuclei sisters]; ovule not increasing in size between pollination and fertilization; pollen grains bicellular at dispersal, germinating in less than 3 hours, siphonogamy, pollen tube unbranched, growing towards the ovule, between cells, growth rate (ca 10-)80-20,000 µm h-1, tube apex of pectins, wall with callose, lumen with callose plugs, penetration of ovules via micropyle [porogamous], whole process takes ca 18 hours, distance to first ovule 1.1-2.1 mm; male gametophytes tricellular, gametes 2, lacking cell walls, ciliae 0, double fertilization +, ovules aborting unless fertilized; fruit indehiscent, P deciduous; mature seed much larger than fertilized ovule, small [<5 mm long], dry [no sarcotesta], exotestal; endosperm +, ?diploid [one polar nucleus + male gamete], cellular, development heteropolar [first division oblique, micropylar end initially with a single large cell, divisions uniseriate, chalazal cell smaller, divisions in several planes], copious, oily and/or proteinaceous, embryo short [<¼ length of seed]; plastid and mitochondrial transmission maternal; Arabidopsis-type telomeres [(TTTAGGG)n]; nuclear genome [2C] (0.57-)1.45(-3.71) [1 pg = 109 base pairs], ??whole nuclear genome duplication [ε/epsilon event]; ndhB gene 21 codons enlarged at the 5' end, single copy of LEAFY and RPB2 gene, knox genes extensively duplicated [A1-A4], AP1/FUL gene, palaeo AP3 and PI genes [paralogous B-class genes] +, with "DEAER" motif, SEP3/LOFSEP and three copies of the PHY gene, [PHYB [PHYA + PHYC]]; chloroplast IR expansions, chlB, -L, -N, trnP-GGG genes 0.
[NYMPHAEALES [AUSTROBAILEYALES [MONOCOTS [[CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]]]]: wood fibres +; axial parenchyma diffuse or diffuse-in-aggregates; pollen monosulcate [anasulcate], tectum reticulate-perforate [here?]; ?genome duplication; "DEAER" motif in AP3 and PI genes lost, gaps in these genes.
[AUSTROBAILEYALES [MONOCOTS [[CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]]]: phloem loading passive, via symplast, plasmodesmata numerous; vessel elements with scalariform perforation plates in primary xylem; essential oils in specialized cells [lamina and P ± pellucid-punctate]; tension wood + [reaction wood: with gelatinous fibres, G-fibres, on adaxial side of branch/stem junction]; anther wall with outer secondary parietal cell layer dividing; tectum reticulate; nucellar cap + [character lost where in eudicots?]; 12BP [4 amino acids] deletion in P1 gene.
[MONOCOTS [[CHLORANTHALES + MAGNOLIIDS] [CERATOPHYLLALES + EUDICOTS]]] / MESANGIOSPERMAE: benzylisoquinoline alkaloids +; sesquiterpene synthase subfamily a [TPS-a] [?level], polyacetate derived anthraquinones + [?level]; outer epidermal walls of root elongation zone with cellulose fibrils oriented transverse to root axis; P more or less whorled, 3-merous [?here]; pollen tube growth intra-gynoecial; extragynoecial compitum 0; carpels plicate [?here]; embryo sac monosporic [spore chalazal], 8-celled, bipolar [Polygonum type], antipodal cells persisting; endosperm triploid.
[CERATOPHYLLALES + EUDICOTS]: ethereal oils 0 [or next node up]; fruit dry [very labile].
EUDICOTS: (Myricetin +), asarone 0 [unknown in some groups, + in some asterids]; root epidermis derived from root cap [?Buxaceae, etc.]; (vessel elements with simple perforation plates in primary xylem); nodes 3:3; stomata anomocytic; flowers (dimerous), cyclic; protandry common; K/outer P members with three traces, ("C" +, with a single trace); A ?, filaments fairly slender, anthers basifixed; microsporogenesis simultaneous, pollen tricolpate, apertures in pairs at six points of the young tetrad [Fischer's rule], cleavage centripetal, wall with endexine; G with complete postgenital fusion, stylulus/style solid [?here], short [<2 x length of ovary]; seed coat?; palaeotetraploidy event.
[PROTEALES [TROCHODENDRALES [BUXALES + CORE EUDICOTS]]]: (axial/receptacular nectary +).
[TROCHODENDRALES [BUXALES + CORE EUDICOTS]]: benzylisoquinoline alkaloids 0; euAP3 + TM6 genes [duplication of paleoAP3 gene: B class], mitochondrial rps2 gene lost.
[BUXALES + CORE EUDICOTS]: mitochondrial rps11 gene lost.
CORE EUDICOTS / GUNNERIDAE: (ellagic and gallic acids +); leaf margins serrate; compitum + [one position]; micropyle?; γ genome duplication [allopolyploidy, 4x x 2x], x = 3 x 7 = 21, 2C genome size (0.79-)1.05(-1.41) pg, PI-dB motif +; small deletion in the 18S ribosomal DNA common.
[ROSIDS ET AL. + ASTERIDS ET AL.] / PENTAPETALAE / [SANTALALES, CARYOPHYLLALES, SAXIFRAGALES, DILLENIALES, VITALES, ROSIDAE, [BERBERIDOPSIDALES + ASTERIDAE]: root apical meristem closed; (cyanogenesis also via [iso]leucine, valine and phenylalanine pathways); flowers rather stereotyped: 5-merous, parts whorled; P = K + C, K enclosing the flower in bud, with three or more traces, odd K adaxial, C with single trace; A = 2x K/C, in two whorls, alternating, (many, but then usually fasciculate and/or centrifugal); pollen tricolporate; G [(3, 4) 5], when 5 opposite K, whorled, placentation axile, style +, stigma not decurrent, compitum + [one position]; endosperm nuclear/coenocytic; fruit dry, dehiscent, loculicidal [when a capsule]; floral nectaries with CRABSCLAW expression, RNase-based gametophytic incompatibility system present.
Phylogeny. Prior to the seventh version of this site asterids were part of a major polytomy that included rosids, Berberidopsidales, Santalales, and Caryophyllales, but then the order of branching immediately below the asterids seemed to be stabilizing, perhaps with a clade [Berberidopsidales [Santalales [Caryophyllales + Asterids]]] while rosid relationships seemed to be [Saxifragales [Vitales + Rosids]]]. However, recent work suggests a polytomy is indeed probably the best way to visualize relationships around here at present. So for further discussion of relationships at the base of asterids and rosids, see the Pentapetalae node.
[SAXIFRAGALES + ROSIDS] / ROSANAE Takhtajan / SUPERROSIDAE: ??
ROSIDS // ROSIDAE: anthers ± dorsifixed, transition to filament narrow, connective thin.
[ROSID I + ROSID II] - Back to Main Tree.
(Mucilage cells with thickened inner periclinal walls and distinct cytoplasm); if nectary +, usu. receptacular; embryo long; chloroplast infA gene defunct, mitochondrial coxII.i3 intron 0.
Age. The age of this node was estimated at (113-)109, 104(-100)Ma by Wikström et al. (2001). H. Wang et al. (2009) suggested an age of (114-)108, 91(-85) Ma or (116-)114, 113(-110) Ma. Magallón and Castillo (2009) suggested ages of ca 107.9 and 108.4 Ma, Moore et al. (2010: 95% HPD) ages of (110-)106(-103) Ma, Bell et al. (2010) ages of (121-)116, 114(-108) Ma, Clarke et al. (2011: Populus + Arabidopsis) an age of (115-)94((-83) Ma, and Sun et al. (2013 ages of 11-108 Ma. In a study using a small sample of nuclear genomes, Argout et al. (2010) obtained an age of ca 90 Ma while the estimate in N. Zhang et al. (2012) is (103-)99(-94) Ma and in Xue et al. (2012) 90.4-89.1 Ma; ages in Schneider et al. (2004) range from about 110-167 Ma, while ages of around 106.3 Ma are provided by Naumann et al. (2013), ca 96 Ma is the age suggested by Sytsma et al. (2014), ca 122Ma by Hohmann et al. (2015: fabids + malvids alone), 116-103 Ma (Zeng et al. 2017: no Zyg.), ca 124 Ma (Foster et al. 2016a: no Zyg.); the oldest, at at around 150 Ma, is that of Z. Wu et al. (2014), while the youngest, at ca 69 Ma, was proposed by Murat et al. (2015b: fab. + mal.).
Poinar et al. (2007, 2008) found a possible rosid fossil from Myanmar/Burma with a floral formula of K 5, C ?, A 10, G [5], styles diverging. It was thought that the rocks in which it was found were 110-100 Ma, but recent estimates are somewhat younger, being Early Cenomanian and (99.4-)98.8(-98.2) Ma (Shi et al. 2012).
Evolution: Divergence & Distribution. Sharkey et al. (2013) suggested that there was a single origin of isoprene emission around here, but this was followed by multiple losses; they link the origin to the high atmospheric CO2 concentrations in the later Cretaceous.
Sun et al. (2019) discuss problems with ascertaining rates and patterns of diversification in the rosids in particular.
Plant-Animal Interactions. Redfern (2011) notes that Cynipinae gall wasps are common on rosids, particularly on Rosaceae and Fagaceae.
Plant-Bacterial/Fungal Associations. Brundrett (2002) suggested that ectomycorrhizae were most common in rosids, rare elsewhere (e.g. Nyctaginaceae), although modified forms also dominate in Ericaceae and Orchidaceae.
Genes & Genomes. Zheng et al. (20113) suggested chromosome numbers of x = 13 or x = 15 for this node.
Chemistry, Morphology, etc.. For the distribution of mucilage cells with thickened inner periclinal walls and distinct cytoplasm ("special mucilage cells"), see Matthews and Endress (2006b), for floral development, see Schönenberger and von Balthazar (2006), and for the distribution of a number of floral features, see Endress and Matthews (2006a).
Phylogeny. See the Dilleniales and Saxifragales pages for further discussion about relationships within Pentapetalae, particularly the basal branches.
ROSID I / FABIDAE / [ZYGOPHYLLALES [COM CLADE + NITROGEN-FIXING CLADE]: endosperm scanty. - Back to Main Tree
Age. Argout et al. (2010) suggested a date for this clade of a mere ca 77 Ma, but this is surely an underestimate. Other ages for this node are (104-)101, 95(-92) Ma (Wikström et al. 2001), (114-)108(-102) and (97-)91(-85) Ma (H. Wang et al. 2009), while ages of (114-)107, 103(-99) Ma were suggested by Bell et al. (2010), around 102.3Ma by Naumann et al. (2013) and ca 113Ma by Tank et al. (2015: Table S1).
Evolution: Divergence & Distribution. H. Wang et al. (2008: penalized likelihood dates) suggested that rapid radiation within Fabidae occurred (114-)108-91(-85) Ma, perhaps a little before that in Malvidae.
Chemistry, Morphology, etc.. Extrafloral nectaries in this clade - perhaps particularly frequent in Malpighiales - commonly are made up of palisade epidermal cells (Zimmermann 1932).
ZYGOPHYLLALES Link - Main Tree.
Harman alkaloids, diversity of lignans and neolignans; mycorrhizae 0; cork cambium deep cortical or pericyclic (superficial); vessel elements with simple perforation plates; rays (predominantly) uniseriate; tension wood?; (stomatal orientation transverse); C clawed; (pollen colpate); style +; micropyle endostomal; seeds ± exotestal; endosperm 0; chloroplast infA gene +. - 2 families, 24 genera, 345 species.
Includes Krameriaceae, Zygophyllaceae.
Note: In all node characterizations, boldface denotes a possible apomorphy, (....) denotes a feature the exact status of which in the clade is uncertain, [....] includes explanatory material; other text lists features found pretty much throughout the clade. However, the precise node to which many characters, particularly the more cryptic ones, should be assigned is unclear. This is partly because homoplasy is very common, in addition, basic information for all too many characters is very incomplete, frequently coming from taxa well embedded in the clade of interest and so making the position of any putative apomorphy uncertain. Then there are the not-so-trivial issues of how character states are delimited and ancestral states are reconstructed (see above).
Age. Ages for crown group Zygophyllales are (88-)79(-70) or (64-)55(-46) Ma (H. Wang et al. 2009: some Bayesian relaxed clock ages up to 102 My). Wikström et al. (2001) suggested an age for the separation of the two families of some (74-)70, 64(-60) Ma, and there are estimates of (88-)70, 65(-45) Ma by Bell et al. (2010), (93.4-)61.9(-29.3) Ma by Naumann et al. (2013), ca 83.5 Ma by Tank et al. (2015: Table S2) and ca 60.9 Ma by Magallón et al. (2015).
Evolution: Divergence & Distribution. Tao et al. (2018) discuss pollen evolution in the order.
Plant-Bacterial/Fungal Associations. Mycorrhizae are usually absent from this clade, perhaps not unexpectedly, given their preference for arid/saline habitats. However, arbuscular mycorrhizae have been reported from roots of Larrea tridentata in the Mojave Desert (Apple et al. 2005); c.f. also Amaranthaceae.
Chemistry, Morphology, etc.. For harman alkaloids, see Kubitzki (2006a); for lignans and neolignans, see Simpson (2006) and Sheahan (2006). Carlquist (2005b) lists several features of wood anatomy that may be synapomorphies for the group.
Phylogeny Zygophyllaceae have often been found to be sister to Krameriaceae, as in Soltis et al. (1998) and Savolainen et al. (2000a).
Classification. The inclusion of Krameriaceae in Zygophyllaceae was initially optional (A.P.G. II 2003), although the two do not have much in common. The narrower circumscription of the families followed here was adopted by A.P.G. III (2009).
Synonymy: Balanitales C. Y. Wu, Krameriales Martius - Zygophyllanae Doweld
KRAMERIACEAE Dumortier, nom. cons. - Kramerbia L. - Back to Zygophyllales
Hemiparasites, shrubs (thorny), (herbs); wood fluorescence?; tannins +; nodes 1:1; petiole bundle (deeply) arcuate; hairs unicellular, thin-walled; leaves amphistomatous [check], stomata usu. paracytic, (anomocytic), transversely oriented; cuticle waxes ± ribbon-like platelets; leaves spiral, (trifoliolate), lamina margins entire, stipules 0; inflorescence racemose, or flowers solitary, pedicels articulated; flower monosymmetric, K (4) 5, C-like adaxially, median member abaxial, C smaller, 3 adaxial C clawed, connate (2(3) when 4-merous), 2 abaxial C not clawed, oil pollination + [epithelial elaiophores]; A 4 (3), (adnate to adaxial C), anthers dehiscing by pores, endothecial cells with thickenings parallel to long axis of cells, filaments often stout; pollen striate; nectary 0; G [2], adaxial member much reduced, style long, stigma small, recessed; ovules 2/carpel, apical, collateral, outer integument 3-6 cells and inner integument 3-5 cells across, suprachalazal zone massive; fruit a nut, with retrorsely barbed spines; seed 1, testa and tegmen ?weakly multiplicative, exotestal cells enlarged, tanniniferous, tegmen to 7 cells thick, both thin, largely disappearing; cotyledons large, cordate/auriculate; n = 6, x = 7, chromosomes 10-24.6 µm long; seedling without root hairs.
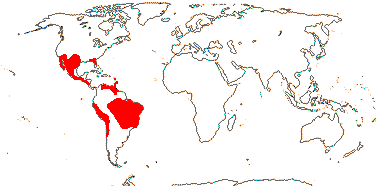
1 [list]/18. S.W. U.S.A. to Chile, the West Indies. Map: from B. B. Simpson et al. (2004). [Photo - Flower © Jim Manhart, Fruit © Dan Nickrent.]
Age. The age of crown group Krameria has been estimated to be (34-)12(-5) Ma (Renner & Schaefer 2010).
Evolution: Ecology & Physiology. Krameria parasitizes a broad variety of plants; it can germinate in the absence of any host (Brokamp 2015). Haustoria develop on the lateral roots and lack phloem, as is common in hemiparasites (Musselman 1976). For more on root parasitism, see Orobanchaceae.
Pollination Biology. Bees (Centris spp.) collect oil from the rather papilionaceous-looking flowers. They hold on to the three adaxial petals and collect oil on their back legs from the epithelial elaiophores of the paired, abaxial petals (Vogel 1974; B. B. Simpson et al. 1977a; Renner & Schaefer 2010; Possobom & Machado 2017a; Tölke et al. 2019 and references). Taxa with 4-merous flowers usually have only two adaxial petals.
Genes & Genomes. The rate of genome evolution here is rather slower than that of other parasitic taxa (Bromham et al. 2013).
Chemistry, Morphology, etc.. The roots have a red phlobaphene pigment; for haustorial development, see Musselman (1975). There are no vessels in the leaves.
Simpson (1982, 2006) discussed the long controversy over the orientation of the flower. The flowers often appear to be inverted or obliquely oriented (see also Kunz 1913; Milby 1971; B. B. Simpson et al. 2004: Fig. 1C; Simpson 2006: Fig. 73), although it is unclear if this is always so (Google Krameria for a complete selection of orientations!); the description above is of an inverted flower. The traces to the sepals, petals and stamens in the flower are all separate; the abaxial petals, which are elaiophores, are densely vascularized (Milby 1971).
For further details, see Simpson (1989, 2006), The Parasitic Plant Collection and Heide-Jørgensen (2008) for general information; Kunz (1913: anatomy and relationships), Carlquist (2005b: wood anatomy), Manning and Stirton (1994: endothecial thickenings), Leinfellner (1971) and especially Bello et al. (2012) for floral morphology and Verkeke (1985: ovule and seed).
Phylogeny. B. B. Simpson et al. (2004) provide a phylogeny of the family, which is made up of two clades in North America and two in South America.
Previous Relationships. Krameriaceae have often been considered to be close to Polygalaceae (Fabales), as by Cronquist (1981).
ZYGOPHYLLACEAE R. Brown, nom. cons. - Back to Zygophyllales
Trees to herbs; mycorrhizae 0; anthraquinones +, guaiacs +, steroid and triterpene saponins, ellagic acid 0, tannins 0 [Zygophyllum]; wood often fluorescing; storying +; pits vestured; nodes often swollen or jointed, 1:1 + split laterals; cortical strands of fibres and sclereids +; petiole bundle annular, with wing bundles; stomata anomocytic; leaves opposite, (odd-) even-pinnate, lamina vernation flat, (secondary veins ± palmate), margins toothed, stipules cauline/1, interpetiolar/(0); ?inflorescence; A obdiplostemonous, adaxial scales at base of A/0; pollen variable; nectary ± annular/lobed (0); G [(2-)5], opposite C, style short to long, stigma as commissural ridges down style, dry or wet; outer integument 2-6 cells across, inner integument 2-4 cells across, endothelium +, (weak nucellar cap +), parietal tissue 1-2(-4) cells across, hypostase +, obturator +; (megaspore mother cells several), embryo sac long; fruit a septicidal capsule or schizocarp, latter opening ventricidally or not; (seed arillate), funicle long [= length of seed]; exotesta often palisade, endotesta crystalliferous, U-lignified or not, endotegmic cells periclinally elongated, lignified; embryo chlorophyllous; x = ?
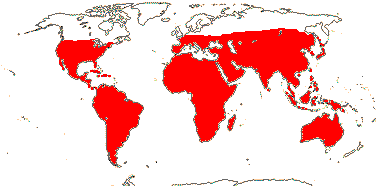
22/325 (280): [list: to subfamilies] - 5 subfamilies below. Dry and warm/cool temperate, also tropical. Map: from Trop. Afr. Fl. Pl. Ecol. Distr. vol. 1 (2003), Beier et al. (2004) and Brummitt (2007), see also Godoy-Bürki et al. (2019).[Photo - Flower, Fruit.]
Age. The crown-group age of Zygophyllaceae is ca 54.8 Ma (S.-D. Wu et al. 2015), (81-)59.9(-38.1) Ma (S.-D. Wu et al. 2018) or ca 65 Ma (Q. Wang et al. 2018).
[Morkillioideae + Tribuloideae]: ?
Age. The age of this clade is around 33 Ma (Q. Wang et al. 2018).
1. Morkillioideae (Engler) Rose & J. H. Painter
Nodes 3:3; leaves spiral, (simple); (flowers/inflorescence opposite leaves), (terminal) (4-merous): ovules 1-several/carpel, epitropous; aril +; (endosperm +, hard); n = ?
3/4. Mexico, Baja California.
2. Tribuloideae (Reichenbach) D. H. Porter ——
Synonymy: Agialidaceae Wettstein, nom. illeg., Balanitaceae M. Roemer nom. cons., Tribulaceae Trautvetter
(Annual herbs); steroidal saponins +; (plant thorny); (nodes 3:3 - Balanites); (C4 photosynthesis); leaves (simple), (anisophyllous), (spiral); adaxial stamen scales usu. 0; (pollen polyporate, metareticulate); (ovary 10-lobed); 1-10 ovules/carpel, (outer integument 4-8 cells across, inner integument 3-6 cells across - Balanites); (fruit a large drupe); n = 6, 10, 12, etc.
6/63: Tribulus (25), Kallstroemia (17). World-wide.
[Seetzenioideae [Larreoideae + Zygophylloideae]]: ?
Age. This clade ia ca 63 Ma (Q. Wang et al. 2018).
3. Seetzenioideae Sheahan & Chase - Seetzenia R. Brown
Prostrate shrublet, ± annual; stem jointed; leaves trifolioliate, stipules interpetiolar; flowers solitary, terminal, (6-merous); K valvate, with thickened basal median strip, C 0; A 5, alternating with K, adaxial stamen scales 0; G alternating with K, styluli usu. 5, radiating, stigmas capitate; ovule 1/carpel, epitropous, micropyle bistomal, outer integument 6-7 cells across, endothelium 0; fruit septicidal, with pyrenes; exotesta not thickened; endosperm +, stony: n + ?
1/2. W. Cape Province, South Africa, N. Africa (the Sahara) to Afghanistan.
[Larreoideae + Zygophylloideae]]: C clawed.
Age. This clade is (66.9-)56.3(-41.1) Ma (S.-D. Wu et al. 2015) or cs 52 Ma (Q. Wang et al. 2018).
4. Larreoideae Sheahan & Chase
Shrub; (CAM photosynthesis + - Bul.); (sieve tube plastids with protein and starch - Larrea); (stems succulent - Bulnesia); adaxial stamen scales common; G stipitate (not); fruit schizocarp, winged or not, 1 seed/loculus; "testa coriaceous", "water storage tissue 0", endosperm +; n = 10, 13.
8/30: Bulnesia (11). Southwest U.S.A., Florida and Mexico to Colombia, the Antilles, extreme south Peru to Chile and Argentina.
Age. The crown-group age of Larreoideae is ca 33 Ma (Q. Wang et al. 2018).
5. Zygophylloideae (R. Brown) Arnott
(Steroidal saponins +); (C4 photosynthesis); leaves (simple), often fleshy, lamina (terete, fleshy), (ephemeral, terete petiole persists), (stipules spinescent), (intrapetiolar); (flowers monosymmetric); K valvate, (C deeply lobed - Augea), initially common A-C primordia [Fagonia]; adaxial stamen scales +/0; (G 3), (pseudo 10-locular), (style ± gynobasic); ovules 1-many/carpel, (straight - Tetraena), (outer integument ca 2 cells across, inner integument ca 2 cells across - Zygophyllum); (fruit loculicidal), (K persistent); (seed arillate), (hilum long); testa often mucilaginous; (endosperm +); 192 BP deletion in trnL UAA intron; n = 7-12.
6/180: Roepera (60), Zygophyllum (50), Tetraena (40), Fagonia (30). Mostly drier areas of the Old World, also S.W. U.S.A. and Chile.
Age. Crown-group Zygophylloideae are (69.4-)38.8, 30.6(-17.1) Ma (Bellstedt et al. (2012), (51.8-)39.8, 37.6(-28.2) Ma (S.-D. Wu et al. 2015) or ca 42 Ma (Q. Wang et al. 2018).
Evolution: Divergence & Distribution. Q. Wang et al. (2018: see S5) give a dated tree for the family; error bars are quite large; for dates, see also S.-D. Wu et al. (2018).
Diversification in Zygophylloideae has been much studied. It is thought that diversification rates increased as climates became cooler and drier (the two are linked) in the middle of the Miocene 18-14 Ma (Q. Wang et al. 2018; S.-D. Wu et al. 2018). The origin of the clade was probably in southern Africa, and there were two movements to Asia and one (by long distance dispersal) to Australia (Bellstedt et al. 2012; S.-D. Wu et al. 2015); Wu et al. (2018) suggest that there have been several dispersal events to other dryland areas, including five alone to the Americas, Morkillioideae and Larreoideae being two of the results. Relationships within Fagonia, widely disjunct between the Old and New Worlds, are quite well understood; in terms of distributions, they can be summarized as [New [Old [Old + New]]], where the two basal clades have but a single species each and the outgroup is Old World (Beier et al. 2004), however, Wu et al. (2015) suggest a somewhat different set of relationships, [New [New [Old + Old]]].
For character evolution, etc., in the southern African Zygophylloideae, see Bellstedt et al. (2008), and for nectary and staminal scale morphology and evolution in Zygophylloideae, see Naghioo et al. (2018).
Ecology & Physiology. Members of Zygophyllaceae are notable components of halophytic vegetation in the Irano-Turanian area and in seasonally dry tropical forests, especially in Central America, but also elsewhere, some species being extremely tolerant of drought and/or of high temperatures (Pennington et al. 2009; Godoy-Bürki et al. 2019; see also articles in Ann. Bot. 115(3). 2015).
Larrea tridentata, the creosote bush, is an important shrub of the deserts of S.W. North America. It is very drought tolerant indeed and can be the only shrub in those deserts; the plant is well known for the strong allelopathic effects that it shows which i.a. leads to the regular spacing of the individuals in the desert (Hierro & Callaway 2021 for references). S. D. Smith et al. (1997 and references) noted that L. tridentata was odd among desert plants in that its photosynthetic rate was little affected by temperature changes and that the plant remained metabolically active during the dry season. Larrea tridentata, as studied in disturbed landscapes in the Southern Chihuahuan Desert, is an important nurse plant, and so affects the recovery of the community as a whole after disturbance and its functioning (Badano et al. 2016). From the literature review in Centenaro et al. (2023: Appendix S1) Larrea tridentata, at ca 11,000 years (11,700 years - Vasek 1980), is the second oldest clonal plant known (Gaylussacia (Ericaceae), at ca 13,000 years, ia the oldest.
C4 photosynthesis is known from genera like Zygophyllum, Tetraena (only T. simplex) and Kallstroemia (Muhaidat et al. 2007; Lauterbach et al. 2016, 2019). In Tetraena there is a switch in C4 types between the flattened cotyledons, which persist for quite a time on the young plant, and the terete leaves that follow (Muhaidat et al. 2018: atriplicoid → kochioid anatomy). Christin et al. (2011b) suggest dates for when the C4 pathway may have been acquired. Lauterbach et al. (2019) note that enlargement of the bundle sheath cells in C3 Tribuloideae may have preceded the development of C4 photosynthesis there, vein density increasing only subsequently; organelles in the bundle sheath cells were close to the bundle itself, as in other C4 plants.
Zygophyllum xanthoxylum has very succulent leaves and tolerates drought; Q. Ma et al. (2024) discuss details of the biosynthesis of cuticular wax and of the use of Na+ and K+ as “cheap" osmolytes, both of them helping the plant withstand the desert conditions in which it lives.
Pollination and Seed Dispersal. In studies of the pollination of the creosote bush, Larrea tridentata, widespread in drier regions in the American southwest, over 120 species of bees were found to be visitors, of which some 22 species of medium-sized to small oligolectic bees were found to use this one species for pollen and nectar, while another 22 species of polylectic bees also regularly visited it (Hurd & Linsley 1975; Danforth et al. 2019; see also Vásquez & Aizen 2004 for such asymmetries in plant-pollinator relationships). These associations seem to have developed with the last 1.6-0.5 Ma (Laport et al. 2012), although other estimates cited by these authors are as much as 8.4 Ma. Zygophyllaceae in the drier areas of southwestern Africa, including Namibia, are much visited by non-Apis bees, which also visit Aizoaceae, Asteraceae and Fabaceae (Kuhlmann & Eardley 2011). Apis mellifera visits Roepera in South Africa for nectar, and sometimes also pollen, depending on the species (Naghiloo et al. 2019). The large basal-adaxial scales that are frequently to be found at the bases of the filaments (not necessarily of all stamens) in Zygophyllaceae at least in some cases protect the nectar from evaporation; these scales may be entire, more or less deeply bifid, or serrate (Naghiloo et al. 2018). Furthermore, the nectary, when present, also varies considerably in its morphology and the number and distribution of the nectarostomata it has (Naghiloo et al. 2018).
A number of species, including those of Zygophyllum, have myrmecochorous seeds (Lengyel et al. 2010), but wind, other forms of animal dispersal and myxospermy also occur (Western 2012).
Plant-Animal Interactions. Ant-associated lycaenid caterpillars are commonly found on Zygophyllaceae (about the second most commonly eaten family, Loranthaceae first), most records being of the southern African Poecilmitis (= Chrysoritis), a copper (Fiedler 1995); for more on lycaenids and plants, see ants and Fabaceae. The two species of Hypermnestra, sister to all other parnassine butterflies (swallowtails) are to be found on Zygophyllaceae, the only butterflies of that group found here (Allio et al. 2020/2021).
Fourteen species of a clade of the cecidomyiid gall formers, the Asphondylia auripila, the creosote gall midges, have diversified on different parts of Larrea tridentata (Joy & Crespi 2007). There are some questions about species limits in Larrea, but L. tridentata, now polyploid, seems to have moved to Mexico—the U.S.A. from South America some time around 8.4-0.5 Ma (Lia et al. 2001; Laport et al. 2014). Although support for the monophyly of the gallers is equivocal, there are a number of monophyletic groups of Asphondylia on L. tridentata - further information on the distribution of the galler would be useful (Joy & Crespi 2007).
Vegetative Variation. Although Zygophyllaceae are quite a small family, vegetative variation is considerable. Lauterbach et al. (2016) discuss variation in southern African Tetraena and Roepera (Zygophylloideae). Leaflets vary from bifacial to terete and unifacial while the C4 T. simplex has terete leaflets and a single central vascular bundle; there is extensive parallel variation in foliar anatomies in the two genera (Lauterbach et al. 2016).
Genes & Genomes. A genome duplication ca 72.8 Ma, the TREIα event, has been associated with crown-group Zygophyllaceae (Landis et al. 2018).
In at least some species of Larrea chloroplasts are inherited paternally (Yang et al. 2000).
Economic Importance. Guaiacum has very hard, self-lubricating heartwood (lignum vitae) that was used in the past to make bearings.
Chemistry, Morphology, etc.. For guaiac, perhaps similar to guaiacol/methoxy phenol/C6H4(OH)(OCH3), see Lambert et al. (2013).
Howard (1970) found no stipules in Balanites, but there are structures in the stipular position there, if minute - colleters? A number of taxa with opposite leaves have split lateral nodes (e.g. Howard 1970), and this may even been the plesiomorphic condition for the family.
There is considerable variation in ovule number, type and arrangement in the family. The style of Zygophyllum is more or less gynobasic.
For a general account of the family, see Sheahan (2006) and G. F. Smith and Figueiredo (2023: succulents), for chemistry, see Hegnauer (1973, 1990), for foliar anatomy, see Sheahan and Cutler (1993), for floral orientation, see Eckert (1966), for androecial development, see Ronse Decraene and Smets (1995c) and for ovule morphology, see Mauritzon (1934b, d), Masand (1963) and Narayana and Rao (1963). Sands (2001) monographed the distinctive Balanites.
Phylogeny. Phylogenetic relationships within the family are fairly well resolved - for clades, see above (Sheahan & Chase 1996, 2000; M. Sun et al. 2016; Q. Wang et al. 2018: S3). However, there are few good characters distinguishing the groups. The very distinctive Balanites is to be included in Tribuloideae (Sheahan & Chase 1996, 2000).
Larreoideae. For details of the relationships of Larrea, see Lia et al. (2001) and especially Godoy-Bürki et al. (2019), in the latter, basal relationships in Larreoideae were unclear, in ITS trees Plectrocarpa being embedded in the new genus Gonopterodendron. Vento et al. (2024: all spp., ITS nuclear rDNA, plastid rbcL, 58 morphological characters) carried out a series of analyses, but did not include ITS in the final analyses, however [Gonopterodendron + Plectrocarpa] came out sister to the subfamily in the ITS analysis and nowhere else, while in the rbcL analysis, Guaiacum was on a very long branch and the outgroup seems to be misplaced; Guaiacum is sister to the rest of the family in the combined analysis. Further work is needed on relationships here.
Tribuloideae. For relationships here, including problems with the circumscription of Tribulus and the possible hybrid origin of C4 Tribulopsis - Kallstroemia x C3 Tribulopsis, see Lauterbach et al. (2019; also Godoy-Bürki et al. 2019). Zygophylloideae. For relationships and morphology here, see Beier et al. (2003). Beier et al. (2004) disentangled relationships within Fagonia, and found the Mexican F. scoparia and the southern European/North African F. cretica to be successively sister to the rest of the genus, although S.-D. Wu et al. (2015) found the latter to be more embedded in the genus. For relationships within the southern African Zygophylloideae, see Bellstedt et al. (2008), also Wu et al. (2015) and Naghiloo et al. (2019), both the whole subfamily, but note that there are some differences in the relationships between the major groups (see also Lauterbach et al. 2016). There are problems with the circumscription of both Fagonia and Zygophyllum in Godoy-Bürki et al. (2019), but this was not the focus of their study.Classification. The subfamilial classification above follows that of Sheahan and Chase (2000). Beier et al. (2003) provide a reclassification of Zygophylloideae, but Bellstedt et al. (2008) suggested that reclassification might be premature given our poor understanding of relationships in the clade. The generic classification in G. F. Smith and Figueiredo (2023) does not follow that used above,
Previous Relationships. Some genera that used to be included in Zygophyllaceae are now in Sapindales-Nitrariaceae.